Deep Learning Enabled Scoring of Pancreatic Neuroendocrine Tumors Based on Cancer Infiltration Patterns
- PMID: 39847242
- PMCID: PMC11757657
- DOI: 10.1007/s12022-025-09846-3
Deep Learning Enabled Scoring of Pancreatic Neuroendocrine Tumors Based on Cancer Infiltration Patterns
Abstract
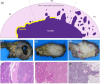
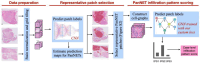
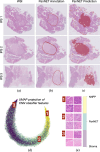
Pancreatic neuroendocrine tumors (PanNETs) are a heterogeneous group of neoplasms that include tumors with different histomorphologic characteristics that can be correlated to sub-categories with different prognoses. In addition to the WHO grading scheme based on tumor proliferative activity, a new parameter based on the scoring of infiltration patterns at the interface of tumor and non-neoplastic parenchyma (tumor-NNP interface) has recently been proposed for PanNET categorization. Despite the known correlations, these categorizations can still be problematic due to the need for human judgment, which may involve intra- and inter-observer variability. Although there is a great need for automated systems working on quantitative metrics to reduce observer variability, there are no such systems for PanNET categorization. Addressing this gap, this study presents a computational pipeline that uses deep learning models to automatically categorize PanNETs for the first time. This pipeline proposes to quantitatively characterize PanNETs by constructing entity-graphs on the cells, and to learn the PanNET categorization using a graph neural network (GNN) trained on these graphs. Different than the previous studies, the proposed model integrates pathology domain knowledge into the GNN construction and training for the purpose of a deeper utilization of the tumor microenvironment and its architectural changes for PanNET categorization. We tested our model on 105 HE stained whole slide images of PanNET tissues. The experiments revealed that this domain knowledge integrated pipeline led to a 76.70% test set F1-score, resulting in significant improvements over its counterparts.
Keywords: Artificial intelligence; Deep learning; Graph neural networks; Infiltration patterns; Pancreatic neuroendocrine tumors.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics Approval: This study was performed in accordance with the Declaration of Helsinki. Competing Interests: The authors declare no competing interests.
Figures

References
-
- Siegel RL, Miller KD, Wagle NS, Jemal A (2024) Cancer statistics, 2024. CA Cancer J Clin. - PubMed
-
- Kos-Kudła B, et al. (2023) European Neuroendocrine Tumour Society (ENETS) 2023 guidance paper for nonfunctioning pancreatic neuroendocrine tumours. J Neuroendocrinol 35(12):e13343. 10.1111/jne.13343 - PubMed
-
- Taskin OC, et al. (2021) Infiltration pattern predicts metastasis and progression better than T-stage and grade in pancreatic neuroendocrine tumors: a proposal for a novel infiltration-based morphologic grading. Mod Pathol 1–9. 10.1038/s41379-021-00838-8 - PubMed
-
- Schiavo Lena M, et al. (2023) Infiltrative growth predicts the risk of recurrence after surgery in well-differentiated non-functioning pancreatic neuroendocrine tumors. Endocr Pathol 34:142–155. 10.1007/s12022-023-09978-2 - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

