Molecular glue for phycobilisome attachment to photosystem II in Synechococcus sp. PCC 7002
- PMID: 39847327
- PMCID: PMC11789067
- DOI: 10.1073/pnas.2415222122
Molecular glue for phycobilisome attachment to photosystem II in Synechococcus sp. PCC 7002
Abstract
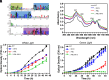
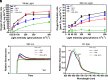
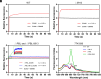
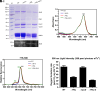
Phycobilisomes (PBS) are the major photosynthetic light-harvesting complexes in cyanobacteria and red algae. While the structures of PBS have been determined in atomic resolutions, how PBS are attached to the reaction centers of photosystems remains less clear. Here, we report that a linker protein (LcpA) is required for the attachment of PBS to photosystem II (PSII) in the cyanobacterium Synechococcus sp. PCC 7002. We also report that the PB-loop of PBS, which is located within the α-APC domain of ApcE, is required for the attachment of PBS to PSII. Deletion of either PB-loop or the gene A0913 led to a decreased rate of photoautotrophic growth under illumination of green light, which is preferentially absorbed by PBS. A double mutant lacking the PB-loop and A0913 (ΔPBL-0913) showed a complete inhibition of O2 evolution under the 590 nm light and could not grow under green light illumination. While assembled PBS could be isolated from ΔPBL-0913, the energy transfer from its PBS to PSII was blocked as measured by fluorescence induction. Photobleaching with intact cells showed that the PBS movement speed in ΔPBL-0913 was 2.5 times as fast as that of the wild type, suggesting that association of its PBS with thylakoids was weakened significantly. The pull-down and coimmunoprecipitation results showed that the LcpA interacts with the CP47 subunit of PSII through its N-terminal region and interacts with ApcB of PBS through its C-terminal α-helix motif. Our results provide insights into the molecular mechanism of PBS-PSII association and shed light on excitation energy transfer from PBS to PSII.
Keywords: cyanobacteria; energy transfer; photosystem II; phycobilisomes.
Conflict of interest statement
Competing interests statement:The authors declare no competing interest.
Figures


Similar articles
-
Phycobilisomes from the mutant cyanobacterium Synechocystis sp. PCC 6803 missing chromophore domain of ApcE.Biochim Biophys Acta Bioenerg. 2018 Apr;1859(4):280-291. doi: 10.1016/j.bbabio.2018.01.003. Epub 2018 Jan 31. Biochim Biophys Acta Bioenerg. 2018. PMID: 29391123
-
Variety in excitation energy transfer processes from phycobilisomes to photosystems I and II.Photosynth Res. 2017 Sep;133(1-3):235-243. doi: 10.1007/s11120-017-0345-3. Epub 2017 Feb 9. Photosynth Res. 2017. PMID: 28185041
-
Altered excitation energy transfer between phycobilisome and photosystems in the absence of ApcG, a small linker peptide, in Synechocystis sp. PCC 6803, a cyanobacterium.Biochim Biophys Acta Bioenerg. 2024 Aug 1;1865(3):149049. doi: 10.1016/j.bbabio.2024.149049. Epub 2024 May 25. Biochim Biophys Acta Bioenerg. 2024. PMID: 38801856
-
ApcD is necessary for efficient energy transfer from phycobilisomes to photosystem I and helps to prevent photoinhibition in the cyanobacterium Synechococcus sp. PCC 7002.Biochim Biophys Acta. 2009 Sep;1787(9):1122-8. doi: 10.1016/j.bbabio.2009.04.007. Epub 2009 May 3. Biochim Biophys Acta. 2009. PMID: 19397890
-
Modulating energy arriving at photochemical reaction centers: orange carotenoid protein-related photoprotection and state transitions.Photosynth Res. 2015 Oct;126(1):3-17. doi: 10.1007/s11120-014-0031-7. Epub 2014 Aug 20. Photosynth Res. 2015. PMID: 25139327 Review.
References
-
- Mahir M., Govindjee V., Semenov A., Nadtochenko A., Primary electron transfer processes in photosynthetic reaction centers from oxygenic organisms. Photosynth Res. 125, 51–63 (2015). - PubMed
-
- Bryant D. A., Canniffe D. P., How nature designs light-harvesting antenna systems: Design principles and functional realization in chlorophototrophic prokaryotes. J Phys. B-At Mol. Opt. 51, 033001 (2018).
MeSH terms
Substances
Grants and funding
- 2022YFF1001700/Ministry of Science and Technology of the People's Republic of China (MOST)
- 32070203/MOST | National Natural Science Foundation of China (NSFC)
- 32270253/MOST | National Natural Science Foundation of China (NSFC)
- 202001539/Qidong-SlS Innovation Funds
- The Fundamental Research Funds for the Central Universities/Peking University (PKU)
LinkOut - more resources
Full Text Sources

