Exploiting YES1-Driven EGFR Expression Improves the Efficacy of EGFR Inhibitors
- PMID: 39847459
- PMCID: PMC12048259
- DOI: 10.1158/1541-7786.MCR-24-0309
Exploiting YES1-Driven EGFR Expression Improves the Efficacy of EGFR Inhibitors
Abstract
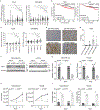
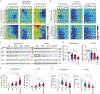
EGFR is a highly expressed driver of many cancers, yet the utility of EGFR inhibitors (EGFRi) is limited to cancers that harbor sensitizing mutations in the EGFR gene because of dose-limiting toxicities. Rather than conventionally blocking the kinase activity of EGFR, we sought to reduce its transcription as an alternative approach to broaden the therapeutic window for EGFR inhibitors targeting wild-type (WT) or mutant EGFR. We found that YES1 is highly expressed in triple-negative breast cancer (TNBC) and drives cell growth by elevating EGFR levels. Mechanistically, YES1 stimulates EGFR expression by signaling to JNK and stabilizing the AP-1 transcription factor c-Jun. This effect extends beyond TNBC as YES1 also sustains EGFR expression in non-small cell lung cancer cells, including those that harbor the EGFR gatekeeper mutation T790M. The novel ability of YES1 to regulate the expression of WT and mutant EGFR mRNA and protein provides a potential therapeutic opportunity of utilizing YES1 blockade to broadly increase the efficacy of EGFR inhibitors. Indeed, we observed synergy within in vitro and in vivo models of TNBC and non-small cell lung cancer, even in the absence of EGFR-activating mutations. Together, these data provide a rationale for blocking YES1 activity as an approach for improving the efficacy of EGFR-targeting drugs in cancers that have generally been refractory to such inhibitors. Implications: YES1 sustains EGFR expression, revealing a therapeutic vulnerability for increasing the efficacy of EGFR inhibitors by lowering the threshold for efficacy in tumors driven by the WT or mutant receptor.
©2025 American Association for Cancer Research.
Conflict of interest statement
Figures



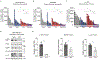
References
MeSH terms
Substances
Grants and funding
- R01CA257502/National Cancer Institute (NCI)
- U54 CA283766/CA/NCI NIH HHS/United States
- T32 CA059366/CA/NCI NIH HHS/United States
- T32GM008056/National Institute of General Medical Sciences (NIGMS)
- T32 GM008056/GM/NIGMS NIH HHS/United States
- 3U54CA283766-02S2/National Cancer Institute (NCI)
- R01 CA257502/CA/NCI NIH HHS/United States
- T32CA059366/National Cancer Institute (NCI)
- R50 CA221675/CA/NCI NIH HHS/United States
- T32GM135081/National Institute of General Medical Sciences (NIGMS)
- T32 GM152319/GM/NIGMS NIH HHS/United States
- R01 CA213843/CA/NCI NIH HHS/United States
- T32GM008803/National Institute of General Medical Sciences (NIGMS)
- T32 GM008803/GM/NIGMS NIH HHS/United States
- T32 GM135081/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

