BAC-browser: the tool for synthetic biology
- PMID: 39849360
- PMCID: PMC11758742
- DOI: 10.1186/s12859-025-06049-9
BAC-browser: the tool for synthetic biology
Abstract
Background: Currently, synthetic genomics is a rapidly developing field. Its main tasks, such as the design of synthetic sequences and the assembly of DNA sequences from synthetic oligonucleotides, require specialized software. In this article, we present a program with a graphical interface that allows non-bioinformatics to perform the tasks needed in synthetic genomics.
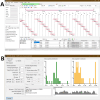
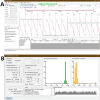
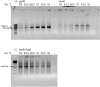
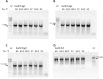
Results: We developed BAC-browser v.2.1. It helps to design nucleotide sequences and features the following tools: generate nucleotide sequence from amino acid sequences using a codon frequency table for a specific organism, as well as visualization of restriction sites, GC composition, GC skew and secondary structure. To assemble DNA sequences, a fragmentation tool was created: regular breakdown into oligonucleotides of a certain length and breakdown into oligonucleotides with thermodynamic alignment. We demonstrate the possibility of DNA fragments assemblies designed in different modes of BAC-browser.
Conclusions: The BAC-browser has a large number of tools for working in the field of systemic genomics and is freely available at the link with a direct link https://sysbiomed.ru/upload/BAC-browser-2.1.zip .
Keywords: Genome browser; LCR assembly; PCR assembly; Sequence editor; Synthetic biology; Synthetic genomics.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethical approval and consent to participate: Not applicable. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures

References
-
- Deaner M, Alper HS. Promoter and terminator discovery and engineering. Adv Biochem Eng Biotechnol. 2016;162:21–44. 10.1007/10_2016_8. - PubMed
-
- Bohrer CH, Fursova NA, Larson DR. Enhancers: a focus on synthetic biology and correlated gene expression. ACS Synth Biol. 2024;13(10):3093–108. 10.1021/ACSSYNBIO.4C00244. - PubMed
-
- Pfleger BF, Pitera DJ, Smolke CD, Keasling JD. Combinatorial engineering of intergenic regions in operons tunes expression of multiple genes. Nat Biotechnol. 2006;24(8):1027–32. 10.1038/NBT1226. - PubMed
-
- Zhang W, Mitchell LA, Bader JS, Boeke JD. Synthetic genomes. Annu Rev Biochem. 2020;89:77–101. 10.1146/ANNUREV-BIOCHEM-013118-110704. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

