De novo genome assembly of the Edwardsiid anthozoan Edwardsia elegans
- PMID: 39849905
- PMCID: PMC12053456
- DOI: 10.1093/g3journal/jkaf011
De novo genome assembly of the Edwardsiid anthozoan Edwardsia elegans
Abstract
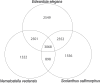
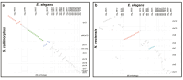
Cnidarians (sea anemones, corals, hydroids, and jellyfish) are a key outgroup for comparisons with bilateral animals to trace the evolution of genomic complexity and diversity within the animal kingdom, as they separated from most other animals 100 s of million years ago. Cnidarians have extensive diversity, yet the paucity of genomic resources limits our ability to compare genomic variation between cnidarian clades and species. Here, we report the genome for Edwardsia elegans, a sea anemone in the most specious genus of the family Edwardsiidae, a phylogenetically important family of sea anemones that contains the model anemone Nematostella vectensis. The E. elegans genome is 396 Mb in length and is predicted to encode approximately 49,000 proteins. We annotated a large conservation of macrosynteny between E. elegans and other Edwardsiidae anemones as well as conservation of both microRNAs and ultra-conserved noncoding elements previously reported in other cnidarians species. We also highlight microsyntenic variation of clustered developmental genes and ancient gene clusters that vary between species of sea anemones, despite previous research showing conservation between cnidarians and bilaterians. Overall, our analysis of the E. elegans genome highlights the importance of using multiple species to represent a taxonomic group for genomic comparisons, where genomic variation can be missed for large and diverse clades.
Keywords: Anthozoa; cnidaria; genome sequencing; genomic diversity; microRNA; synteny.
© The Author(s) 2025. Published by Oxford University Press on behalf of The Genetics Society of America.
Conflict of interest statement
Conflicts of interest: The authors declare no conflict of interest.
Figures




Similar articles
-
Nematostella vectensis exemplifies the exceptional expansion and diversity of opsins in the eyeless Hexacorallia.Evodevo. 2023 Sep 21;14(1):14. doi: 10.1186/s13227-023-00218-8. Evodevo. 2023. PMID: 37735470 Free PMC article.
-
Two species of Edwardsia having gigantic nematocysts, E. aff. tuberculata br />and E. alternobomen sp. nov. (Cnidaria; Anthozoa; Actiniaria; Edwardsiidae) from Japan.Zootaxa. 2019 Aug 29;4661(3):zootaxa.4661.3.7. doi: 10.11646/zootaxa.4661.3.7. Zootaxa. 2019. PMID: 31716701
-
The genome of the deep-sea anemone Actinernus sp. contains a mega-array of ANTP-class homeobox genes.Proc Biol Sci. 2023 Oct 25;290(2009):20231563. doi: 10.1098/rspb.2023.1563. Epub 2023 Oct 25. Proc Biol Sci. 2023. PMID: 37876192 Free PMC article.
-
Recent advances in genomics and transcriptomics of cnidarians.Mar Genomics. 2015 Dec;24 Pt 2:131-8. doi: 10.1016/j.margen.2015.09.007. Epub 2015 Oct 1. Mar Genomics. 2015. PMID: 26421490 Review.
-
Rising starlet: the starlet sea anemone, Nematostella vectensis.Bioessays. 2005 Feb;27(2):211-21. doi: 10.1002/bies.20181. Bioessays. 2005. PMID: 15666346 Review.
Cited by
-
A Network Approach to White Band Disease Challenged Staghorn Coral Acropora cervicornismicroRNAs and Their Targets.Ecol Evol. 2025 Apr 25;15(4):e71351. doi: 10.1002/ece3.71351. eCollection 2025 Apr. Ecol Evol. 2025. PMID: 40290387 Free PMC article.
-
Repeatome diversity in sea anemone genomics (Cnidaria: Actiniaria) based on the Actiniaria-REPlib library.BMC Genomics. 2025 May 13;26(1):473. doi: 10.1186/s12864-025-11591-0. BMC Genomics. 2025. PMID: 40361000 Free PMC article.
References
-
- Al-Shaer L, Havrilak J, Layden MJ. 2021. Nematostella vectensis as a model system. In: Handbook of marine model organisms in experimental biology. CRC Press. p. 107–128.
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
