Comprehensive Analysis of the Fourteen Complete Genome Sequences of Buchnera aphidicola Isolated from Aphis Species
- PMID: 39849922
- PMCID: PMC11813355
- DOI: 10.4014/jmb.2409.09004
Comprehensive Analysis of the Fourteen Complete Genome Sequences of Buchnera aphidicola Isolated from Aphis Species
Abstract
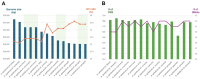
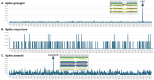
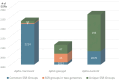
Endosymbionts are important for insect species as they provide essential substances to the host. Due to the technical advance of NGS technology and de novo assemblers, many endosymbionts bacterial genomes are available now. Here, we analysed fourteen endosymbiont bacterial genomes of Aphis genius, one of notorious pest species. Fourteen genomes displayed the length between 628,098 bp to 634,931 bp; GC ratio was from 24.2 % to 25.6 % with no structural variation found. The nucleotide diversity distribution across the 14 endosymbiont genomes revealed three distinct regions, each separated by varying levels of nucleotide diversity. Intraspecific variations identified from endosymbiont bacterial genomes of the same host species revealed numbers of SNPs ranging from 31 (0.0049%) to 1,652 (0.26%) and those of INDELs ranging from 7 (21 bp; 0.0033%) to 104 (285 bp; 0.0045%). 250 unique SSRs, 28 different common SSR groups, and one different SSR group in two genomes were identified and used as a potential molecular marker to distinguish intraspecific population. Phylogenetic analysis further showed congruence between the endosymbiont bacterial genomes and the host species phylogeny, except Aphis nasturtii, Aphis helianth, and Aphis auranti, which require additional endosymbiont genomes for clarification. This comparative analysis result could serve as a cornerstone for understanding the relationship between host and endosymbiont species from a genomic perspective.
Keywords: Aphis; Buchnera aphidicola; endosymbiont bacterial genome; intraspecific variations; nucleotide diversity; phylogenetic analysis; simple sequence repeats.
Conflict of interest statement
The authors have no financial conflicts of interest to declare.
Figures






References
-
- Vega FE, Dowd PF. The role of yeasts as insect endosymbionts. Insect-Fungal Associations: Ecology and Evolution. Oxford University Press, New York; 2005. pp. 211–243.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

