Combination adjuvant improves influenza virus immunity by downregulation of immune homeostasis genes in lymphocytes
- PMID: 39849993
- PMCID: PMC11841980
- DOI: 10.1093/immhor/vlae007
Combination adjuvant improves influenza virus immunity by downregulation of immune homeostasis genes in lymphocytes
Abstract
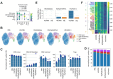
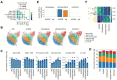
Adjuvants play a central role in enhancing the immunogenicity of otherwise poorly immunogenic vaccine antigens. Combining adjuvants has the potential to enhance vaccine immunogenicity compared with single adjuvants, although the cellular and molecular mechanisms of combination adjuvants are not well understood. Using the influenza virus hemagglutinin H5 antigen, we define the immunological landscape of combining CpG and MPLA (TLR-9 and TLR-4 agonists, respectively) with a squalene nanoemulsion (AddaVax) using immunologic and transcriptomic profiling. Mice immunized and boosted with recombinant H5 in AddaVax, CpG+MPLA, or AddaVax plus CpG+MPLA (IVAX-1) produced comparable levels of neutralizing antibodies and were equally well protected against the H5N1 challenge. However, after challenge with H5N1 virus, H5/IVAX-1-immunized mice had 100- to 300-fold lower virus lung titers than mice receiving H5 in AddaVax or CpG+MPLA separately. Consistent with enhanced viral clearance, unsupervised expression analysis of draining lymph node cells revealed the combination adjuvant IVAX-1 significantly downregulated immune homeostasis genes, and induced higher numbers of antibody-producing plasmablasts than either AddaVax or CpG+MPLA. IVAX-1 was also more effective after single-dose administration than either AddaVax or CpG+MPLA. These data reveal a novel molecular framework for understanding the mechanisms of combination adjuvants, such as IVAX-1, and highlight their potential for the development of more effective vaccines against respiratory viruses.
Keywords: combination adjuvant; influenza; lymphocytes; single-cell transcriptomics; synergy.
© The Author(s) 2025. Published by Oxford University Press on behalf of The American Association of Immunologists.
Conflict of interest statement
None declared.
Figures



Similar articles
-
Administration of Multivalent Influenza Virus Recombinant Hemagglutinin Vaccine in Combination-Adjuvant Elicits Broad Reactivity Beyond the Vaccine Components.Front Immunol. 2021 Jul 14;12:692151. doi: 10.3389/fimmu.2021.692151. eCollection 2021. Front Immunol. 2021. PMID: 34335601 Free PMC article.
-
Recombinant hemagglutinin produced from Chinese Hamster Ovary (CHO) stable cell clones and a PELC/CpG combination adjuvant for H7N9 subunit vaccine development.Vaccine. 2019 Nov 8;37(47):6933-6941. doi: 10.1016/j.vaccine.2019.02.040. Epub 2019 Aug 2. Vaccine. 2019. PMID: 31383491 Free PMC article.
-
Recombinant H5 hemagglutinin adjuvanted with nanoemulsion protects ferrets against pathogenic avian influenza virus challenge.Vaccine. 2019 Mar 14;37(12):1591-1600. doi: 10.1016/j.vaccine.2019.02.002. Epub 2019 Feb 19. Vaccine. 2019. PMID: 30795941
-
Magnitude and breadth of antibody cross-reactivity induced by recombinant influenza hemagglutinin trimer vaccine is enhanced by combination adjuvants.Sci Rep. 2022 Jun 2;12(1):9198. doi: 10.1038/s41598-022-12727-y. Sci Rep. 2022. PMID: 35654904 Free PMC article.
-
The ABC of clinical and experimental adjuvants--a brief overview.Immunol Lett. 2010 Jan 18;128(1):29-35. doi: 10.1016/j.imlet.2009.10.005. Epub 2009 Nov 4. Immunol Lett. 2010. PMID: 19895847 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
- University of California, Irvine
- R01 AI168063/AI/NIAID NIH HHS/United States
- National Institute of Allergy and Infectious Diseases
- U01AI160397/Molecular Mechanisms of Combination Adjuvants
- 594598/Simons Foundation
- 1S10OD010794-01/Genomics Research and Technology Hub
- U01 AI160397/AI/NIAID NIH HHS/United States
- HDTRA-1-18-0036/Defense Threat Reduction Agency
- R01AI168063/AI/NIAID NIH HHS/United States
- DMS1763272/National Science Foundation
- P30 CA062203/CA/NCI NIH HHS/United States
- P30AR075047/UCI Skin Biology Resource Based Center
- S10 OD021718/OD/NIH HHS/United States
- S10 OD010794/OD/NIH HHS/United States
- NIH
- P30 AR075047/AR/NIAMS NIH HHS/United States
- Genomics Research and Technology Hub
- P30CA-062203/Comprehensive Cancer Center
LinkOut - more resources
Full Text Sources
Medical

