Somatic mutations and DNA methylation identify a subgroup of poor prognosis within lower-risk myelodysplastic syndromes
- PMID: 39850648
- PMCID: PMC11754767
- DOI: 10.1002/hem3.70073
Somatic mutations and DNA methylation identify a subgroup of poor prognosis within lower-risk myelodysplastic syndromes
Abstract
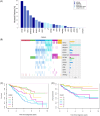
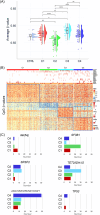
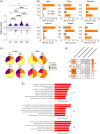
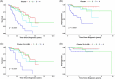
Lower risk (LR) myelodysplastic syndromes (MDS) are heterogeneous hematopoietic stem and progenitor disorders caused by the accumulation of somatic mutations in various genes including epigenetic regulators that may produce convergent DNA methylation patterns driving specific gene expression profiles. The integration of genomic, epigenomic, and transcriptomic profiling has the potential to spotlight distinct LR-MDS categories on the basis of pathophysiological mechanisms. We performed a comprehensive study of somatic mutations and DNA methylation in a large and clinically well-annotated cohort of treatment-naive patients with LR-MDS at diagnosis from the EUMDS registry (ClinicalTrials.gov.NCT00600860). Unsupervised clustering analyses identified six clusters based on genetic profiling that concentrate into four clusters on the basis of genome-wide methylation profiling with significant overlap between the two clustering modes. The four methylation clusters showed distinct clinical and genetic features and distinct methylation landscape. All clusters shared hypermethylated enhancers enriched in binding motifs for ETS and bZIP (C/EBP) transcription factor families, involved in the regulation of myeloid cell differentiation. By contrast, one cluster gathering patients with early leukemic evolution exhibited a specific pattern of hypermethylated promoters and, distinctly from other clusters, the upregulation of AP-1 complex members FOS/FOSL2 together with the absence of hypermethylation of their binding motif at target gene enhancers, which is of relevance for leukemic initiation. Among MDS patients with lower-risk IPSS-M, this cluster displayed a significantly inferior overall survival (p < 0.0001). Our study showed that genetic and DNA methylation features of LR-MDS at early stages may refine risk stratification, therefore offering the frame for a precocious therapeutic intervention.
© 2025 The Author(s). HemaSphere published by John Wiley & Sons Ltd on behalf of European Hematology Association.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- de Swart L, Smith A, Johnston TW, et al. Validation of the revised international prognostic scoring system (IPSS‐R) in patients with lower‐risk myelodysplastic syndromes: a report from the prospective European LeukaemiaNet MDS (EUMDS) registry. Br J Haematol. 2015;170(3):372‐383. 10.1111/bjh.13450 - DOI - PubMed
Associated data
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous
