Antibiotic-mediated dysbiosis leads to activation of inflammatory pathways
- PMID: 39850904
- PMCID: PMC11754057
- DOI: 10.3389/fimmu.2024.1493991
Antibiotic-mediated dysbiosis leads to activation of inflammatory pathways
Abstract
Introduction: The gut microbiota plays a pivotal role in influencing host health, through the production of metabolites and other key signalling molecules. While the impact of specific metabolites or taxa on host cells is well-documented, the broader impact of a disrupted microbiota on immune homeostasis is less understood, which is particularly important in the context of the increasing overuse of antibiotics.
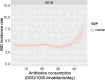
Methods: Female C57BL/6 mice were gavaged twice daily for four weeks with Vancomycin, Polymyxin B, or PBS (control). Caecal microbiota composition was assessed via 16S rRNA sequencing and caecal metabolites were quantified with NMR spectroscopy. Immune profiles of spleen and mesenteric lymph nodes (MLNs) were assessed by flow cytometry, and splenocytes assessed for ex vivo cytokine production. A generalised additive model approach was used to examine the relationship between global antibiotic consumption and IBD incidence.
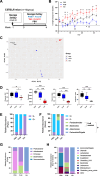
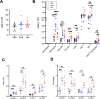
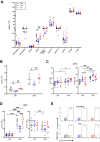
Results: Antibiotics significantly altered gut microbiota composition, reducing alpha-diversity. Acetate and butyrate were significantly reduced in antibiotic groups, while propionate and succinate increased in Vancomycin and PmB-treated mice, respectively. The MLNs and spleen showed changes only to DC numbers. Splenocytes from antibiotic-treated mice stimulated ex vivo exhibited increased production of TNF. Epidemiological analysis revealed a positive correlation between global antibiotic consumption and IBD incidence.
Discussion: Our findings demonstrate that antibiotic-mediated dysbiosis results in significantly altered short-chain fatty acid levels but immune homeostasis in spleen and MLNs at steady state is mostly preserved. Non-specific activation of splenocytes ex vivo, however, revealed mice with perturbed microbiota had significantly elevated production of TNF. Thus, this highlights antibiotic-mediated disruption of the gut microbiota may program the host towards dysregulated immune responses, predisposing to the development of TNF-associated autoimmune or chronic inflammatory disease.
Keywords: IBD; Polymyxin B; TNF; antibiotics; autoimmunity; dysbiosis; gut microbiota; vancomycin.
Copyright © 2025 Taitz, Tan, Ni, Potier-Villette, Grau, Nanan and Macia.
Conflict of interest statement
LM is a current employee of the Translational Science Hub Global Sanofi Vaccines R&D Brisbane, Australia. Her contribution to this manuscript was when she was an employee of the University of Sydney. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Figures


Similar articles
-
Antibiotic-associated dysbiosis affects the ability of the gut microbiota to control intestinal inflammation upon fecal microbiota transplantation in experimental colitis models.Microbiome. 2021 Feb 6;9(1):39. doi: 10.1186/s40168-020-00991-x. Microbiome. 2021. PMID: 33549144 Free PMC article.
-
Alterations in gut microbiota and inflammatory cytokines after administration of antibiotics in mice.Microbiol Spectr. 2024 Aug 6;12(8):e0309523. doi: 10.1128/spectrum.03095-23. Epub 2024 Jun 20. Microbiol Spectr. 2024. PMID: 38899904 Free PMC article.
-
Alteration of Colonic Mucosal Permeability during Antibiotic-Induced Dysbiosis.Int J Mol Sci. 2020 Aug 25;21(17):6108. doi: 10.3390/ijms21176108. Int J Mol Sci. 2020. PMID: 32854266 Free PMC article.
-
Links Between Inflammatory Bowel Disease and Chronic Obstructive Pulmonary Disease.Front Immunol. 2020 Sep 11;11:2144. doi: 10.3389/fimmu.2020.02144. eCollection 2020. Front Immunol. 2020. PMID: 33042125 Free PMC article. Review.
-
Connecting the immune system, systemic chronic inflammation and the gut microbiome: The role of sex.J Autoimmun. 2018 Aug;92:12-34. doi: 10.1016/j.jaut.2018.05.008. Epub 2018 Jun 1. J Autoimmun. 2018. PMID: 29861127 Review.
Cited by
-
Absence of gut microbiota restoration following meropenem treatment in a systemic infection model.NPJ Antimicrob Resist. 2025 Jun 30;3(1):60. doi: 10.1038/s44259-025-00129-9. NPJ Antimicrob Resist. 2025. PMID: 40588535 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

