Identification of serum tRNA-derived small RNAs biosignature for diagnosis of tuberculosis
- PMID: 39851057
- PMCID: PMC11803760
- DOI: 10.1080/22221751.2025.2459132
Identification of serum tRNA-derived small RNAs biosignature for diagnosis of tuberculosis
Abstract
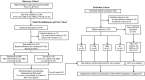
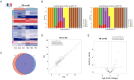
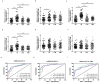
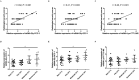
The tRNA-derived small RNAs (tsRNAs) are a new class of non coding RNAs, which are stable in body fluids and can be used as potential biomarkers for disease diagnosis. However, the exact value of tsRNAs in the diagnosis of tuberculosis (TB) is still unclear. The objective of the present study was to evaluate the performance of the serum tsRNAs biosignature to distinguish between active TB, healthy controls, latent TB infection, and other respiratory diseases. The differential expression profiles of tsRNAs in serum from active TB patients and healthy controls were analyzed by high-throughput sequencing. A total of 905 subjects were prospectively recruited for our study from three different cohorts. Levels of tsRNA-Gly-CCC-2, tsRNA-Gly-GCC-1, and tsRNA-Lys-CTT-2-M2 were significantly elevated in the serum of TB patients compared to non-TB individuals, showing a correlation with lung injury severity and acid-fast bacilli grades in TB patients. The accuracy of the three-tsRNA biosignature for TB diagnosis was evaluated in the training (n = 289), test (n = 124), and prediction (n = 292) groups. By utilizing cross-validation with a random forest algorithm approach, the training cohort achieved a sensitivity of 100% and specificity of 100%. The test cohort exhibited a sensitivity of 75.8% and a specificity of 91.2%. Within the prediction group, the sensitivity and specificity were 73.1% and 92.5%, respectively. The three-tsRNA biosignature generally decreased within 3 months of treatment and then remained stable. In conclusion, the three-tsRNA biosignature might serve as biomarker to diagnose TB and to monitor the effectiveness of treatment in a high-burden TB clinical setting.
Keywords: Tuberculosis; biomarker; biosignature; diagnosis; transfer RNA-derived small RNAs.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures

Similar articles
-
Transfer RNA-derived small RNAs (tsRNAs) sequencing revealed a differential expression landscape of tsRNAs between glioblastoma and low-grade glioma.Gene. 2023 Mar 1;855:147114. doi: 10.1016/j.gene.2022.147114. Epub 2022 Dec 13. Gene. 2023. PMID: 36526122
-
A Novel Serum tsRNA for Diagnosis and Prediction of Nephritis in SLE.Front Immunol. 2021 Nov 11;12:735105. doi: 10.3389/fimmu.2021.735105. eCollection 2021. Front Immunol. 2021. PMID: 34867955 Free PMC article.
-
Identification of eight-protein biosignature for diagnosis of tuberculosis.Thorax. 2020 Jul;75(7):576-583. doi: 10.1136/thoraxjnl-2018-213021. Epub 2020 Mar 22. Thorax. 2020. PMID: 32201389 Free PMC article.
-
Unveiling the role of tRNA-derived small RNAs in MAPK signaling pathway: implications for cancer and beyond.Front Genet. 2024 Mar 26;15:1346852. doi: 10.3389/fgene.2024.1346852. eCollection 2024. Front Genet. 2024. PMID: 38596214 Free PMC article. Review.
-
Novel insights into the roles of tRNA-derived small RNAs.RNA Biol. 2021 Dec;18(12):2157-2167. doi: 10.1080/15476286.2021.1922009. Epub 2021 May 17. RNA Biol. 2021. PMID: 33998370 Free PMC article. Review.
Cited by
-
RNA Polymerase III-Transcribed RNAs in Health and Disease: Mechanisms, Dysfunction, and Future Directions.Int J Mol Sci. 2025 Jun 18;26(12):5852. doi: 10.3390/ijms26125852. Int J Mol Sci. 2025. PMID: 40565315 Free PMC article. Review.
References
-
- World Health Organization . Global tuberculosis report 2023. Geneva: World Health Organization; 2023.
-
- World Health Organization . High-priority target product profiles for new tuberculosis diagnostics: report of a consensus meeting. Geneva: World Health Organization; 2014.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
