The MCPH7 Gene Product STIL Is Essential for Dendritic Spine Formation
- PMID: 39851490
- PMCID: PMC11764357
- DOI: 10.3390/cells14020062
The MCPH7 Gene Product STIL Is Essential for Dendritic Spine Formation
Abstract
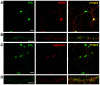
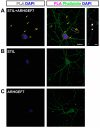
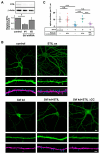
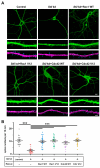
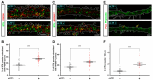
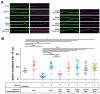
Dendritic spine formation/maintenance is highly dependent on actin cytoskeletal dynamics, which is regulated by small GTPases Rac1 and Cdc42 through their downstream p21-activated kinase/LIM-kinase-I/cofilin pathway. ARHGEF7, also known as ß-PIX, is a guanine nucleotide exchange factor for Rac1 and Cdc42, thereby activating Rac1/Cdc42 and the downstream pathway, leading to the upregulation of spine formation/maintenance. We found that STIL, one of the primary microcephaly gene products, is associated with ARHGEF7 in dendritic spines and that knockdown of Stil resulted in a significant reduction in dendritic spines in neurons both in vitro and in vivo. Rescue experiments indicated that the STIL requirement for spine formation/maintenance depended on its coiled coil domain that mediates the association with ARHGEF7. The overexpression of Rac1/Cdc42 compensated for the spine reduction caused by STIL knockdown. FRET experiments showed that Rac activation is impaired in STIL knockdown neurons. Chemical long-term potentiation, which triggers Rac activation, promoted STIL accumulation in the spine and its association with ARHGEF7. The dynamics of these proteins further supported their coordinated involvement in spine formation/maintenance. Based on these findings, we concluded that the centrosomal protein STIL is a novel regulatory factor essential for spine formation/maintenance by activating Rac and its downstream pathway, possibly through the association with ARHGEF7.
Keywords: ARHGEF7; Cdc42; MCPH7; Rac1; STIL; dendritic spine.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
Indispensable role of STIL in the regulation of cancer cell motility through the lamellipodial accumulation of ARHGEF7-PAK1 complex.Oncogene. 2020 Feb;39(9):1931-1943. doi: 10.1038/s41388-019-1115-9. Epub 2019 Nov 21. Oncogene. 2020. PMID: 31754215
-
The Shank family of postsynaptic density proteins interacts with and promotes synaptic accumulation of the beta PIX guanine nucleotide exchange factor for Rac1 and Cdc42.J Biol Chem. 2003 May 23;278(21):19220-9. doi: 10.1074/jbc.M301052200. Epub 2003 Mar 7. J Biol Chem. 2003. PMID: 12626503
-
GEFT, a Rho family guanine nucleotide exchange factor, regulates neurite outgrowth and dendritic spine formation.J Biol Chem. 2004 Oct 29;279(44):45824-32. doi: 10.1074/jbc.M406216200. Epub 2004 Aug 17. J Biol Chem. 2004. PMID: 15322108
-
Expanding functions of GIT Arf GTPase-activating proteins, PIX Rho guanine nucleotide exchange factors and GIT-PIX complexes.J Cell Sci. 2016 May 15;129(10):1963-74. doi: 10.1242/jcs.179465. J Cell Sci. 2016. PMID: 27182061 Free PMC article. Review.
-
Stress and trauma: BDNF control of dendritic-spine formation and regression.Prog Neurobiol. 2014 Jan;112:80-99. doi: 10.1016/j.pneurobio.2013.10.005. Epub 2013 Nov 6. Prog Neurobiol. 2014. PMID: 24211850 Review.
Cited by
-
Arhgef7 as a key target for enriched environment rescuing spatial cognitive deficits and anxiety-like behaviors in a mouse model of Alzheimer's disease following early social isolation.Alzheimers Res Ther. 2025 Jul 1;17(1):143. doi: 10.1186/s13195-025-01797-5. Alzheimers Res Ther. 2025. PMID: 40598577 Free PMC article.
References
-
- Ramón y Cajal S. Neue darstellung vom histologischen bau des centralnervensystem. Arch. Anat. Entwick. 1893;3:319–428.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

