Transcriptomic and Functional Landscape of Adult Human Spinal Cord NSPCs Compared to iPSC-Derived Neural Progenitor Cells
- PMID: 39851491
- PMCID: PMC11763936
- DOI: 10.3390/cells14020064
Transcriptomic and Functional Landscape of Adult Human Spinal Cord NSPCs Compared to iPSC-Derived Neural Progenitor Cells
Abstract
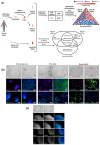
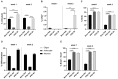
The adult human spinal cord harbors diverse populations of neural stem/progenitor cells (NSPCs) essential for neuroregeneration and central nervous system repair. While induced pluripotent stem cell (iPSC)-derived NSPCs offer significant therapeutic potential, understanding their molecular and functional alignment with bona fide spinal cord NSPCs is crucial for developing autologous cell therapies that enhance spinal cord regeneration and minimize immune rejection. In this study, we present the first direct transcriptomic and functional comparison of syngeneic adult human NSPC populations, including bona fide spinal cord NSPCs and iPSC-derived NSPCs regionalized to the spinal cord (iPSC-SC) and forebrain (iPSC-Br). RNA sequencing analysis revealed distinct transcriptomic profiles and functional disparities among NSPC types. iPSC-Br NSPCs exhibited a close resemblance to bona fide spinal cord NSPCs, characterized by enriched expression of neurogenesis, axon guidance, synaptic signaling, and voltage-gated calcium channel activity pathways. Conversely, iPSC-SC NSPCs displayed significant heterogeneity, suboptimal regional specification, and elevated expression of neural crest and immune response-associated genes. Functional assays corroborated the transcriptomic findings, demonstrating superior neurogenic potential in iPSC-Br NSPCs. Additionally, we assessed donor-specific influences on NSPC behavior by analyzing gene expression and differentiation outcomes across syngeneic populations from multiple individuals. Donor-specific factors significantly modulated transcriptomic profiles, with notable variability in the alignment of iPSC-derived NSPCs to bona fide spinal cord NSPCs. Enrichment of pathways related to neurogenesis, axon guidance, and synaptic signaling varied across donors, highlighting the impact of genetic and epigenetic individuality on NSPC behavior.
Keywords: cellular heterogeneity; differentiation potential; induced pluripotent stem cells; neural stem/progenitor cells; neurogenesis; patient-specific variability; proliferation dynamics; regenerative medicine; spinal cord injury; transcriptomics.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures



References
-
- Saremi J., Mahmoodi N., Rasouli M., Ranjbar F.E., Mazaheri E.L., Akbari M., Hasanzadeh E., Azami M. Advanced approaches to regenerate spinal cord injury: The development of cell and tissue engineering therapy and combinational treatments. Biomed. Pharmacother. 2022;146:112529. doi: 10.1016/j.biopha.2021.112529. - DOI - PubMed
-
- Tetzlaff W., Okon E.B., Karimi-Abdolrezaee S., Hill C.E., Sparling J.S., Plemel J.R., Plunet W.T., Tsai E.C., Baptiste D., Smithson L.J., et al. A systematic review of cellular transplantation therapies for spinal cord injury. J. Neurotrauma. 2011;28:1611–1682. doi: 10.1089/neu.2009.1177. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

