Ubiquitination Enzymes in Cancer, Cancer Immune Evasion, and Potential Therapeutic Opportunities
- PMID: 39851497
- PMCID: PMC11763706
- DOI: 10.3390/cells14020069
Ubiquitination Enzymes in Cancer, Cancer Immune Evasion, and Potential Therapeutic Opportunities
Abstract
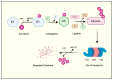
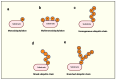
Ubiquitination is cells' second most abundant posttranslational protein modification after phosphorylation. The ubiquitin-proteasome system (UPS) is critical in maintaining essential life processes such as cell cycle control, DNA damage repair, and apoptosis. Mutations in ubiquitination pathway genes are strongly linked to the development and spread of multiple cancers since several of the UPS family members possess oncogenic or tumor suppressor activities. This comprehensive review delves into understanding the ubiquitin code, shedding light on its role in cancer cell biology and immune evasion. Furthermore, we highlighted recent advances in the field for targeting the UPS pathway members for effective therapeutic intervention against human cancers. We also discussed the recent update on small-molecule inhibitors and PROTACs and their progress in preclinical and clinical trials.
Keywords: E3 ligases; PROTACs; cancer; cell cycle; immune evasion; oncogenes; tumor suppressor; ubiquitination.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
Similar articles
-
Ubiquitination and deubiquitination: Implications on cancer therapy.Biochim Biophys Acta Gene Regul Mech. 2023 Dec;1866(4):194979. doi: 10.1016/j.bbagrm.2023.194979. Epub 2023 Aug 24. Biochim Biophys Acta Gene Regul Mech. 2023. PMID: 37633647 Review.
-
Expanding the ubiquitin code in pancreatic cancer.Biochim Biophys Acta Mol Basis Dis. 2024 Jan;1870(1):166884. doi: 10.1016/j.bbadis.2023.166884. Epub 2023 Sep 12. Biochim Biophys Acta Mol Basis Dis. 2024. PMID: 37704111 Review.
-
Ubiquitination and deubiquitination in cancer: from mechanisms to novel therapeutic approaches.Mol Cancer. 2024 Jul 25;23(1):148. doi: 10.1186/s12943-024-02046-3. Mol Cancer. 2024. PMID: 39048965 Free PMC article. Review.
-
The role of the ubiquitin-proteasome pathway in cancer development and treatment.Front Biosci (Landmark Ed). 2014 Jun 1;19(6):886-95. doi: 10.2741/4254. Front Biosci (Landmark Ed). 2014. PMID: 24896323 Review.
-
Pharmacological Modulation of Ubiquitin-Proteasome Pathways in Oncogenic Signaling.Int J Mol Sci. 2021 Nov 4;22(21):11971. doi: 10.3390/ijms222111971. Int J Mol Sci. 2021. PMID: 34769401 Free PMC article. Review.
Cited by
-
The Emerging Role and Mechanism of E2/E3 Hybrid Enzyme UBE2O in Human Diseases.Biomedicines. 2025 Apr 29;13(5):1082. doi: 10.3390/biomedicines13051082. Biomedicines. 2025. PMID: 40426910 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

