Exploiting Cancer Dormancy Signaling Mechanisms in Epithelial Ovarian Cancer Through Spheroid and Organoid Analysis
- PMID: 39851561
- PMCID: PMC11764263
- DOI: 10.3390/cells14020133
Exploiting Cancer Dormancy Signaling Mechanisms in Epithelial Ovarian Cancer Through Spheroid and Organoid Analysis
Abstract
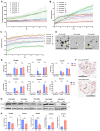
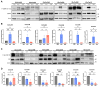
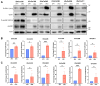
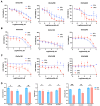
Epithelial ovarian cancer (EOC) exhibits a unique mode of metastasis, involving spheroid formation in the peritoneum. Our research on EOC spheroid cell biology has provided valuable insights into the signaling plasticity associated with metastasis. We speculate that EOC cells modify their biology between tumour and spheroid states during cancer dormancy, although the specific mechanisms underlying this transition remain unknown. Here, we present novel findings from direct comparisons between cultured EOC spheroids and organoids. Our results indicated that AMP-activated protein kinase (AMPK) activity was significantly upregulated and protein kinase B (Akt) was downregulated in EOC spheroids compared to organoids, suggesting a clear differential phenotype. Through RNA sequencing analysis, we further supported these phenotypic differences and highlighted the significance of cell cycle regulation in organoids. By inhibiting the G2/M checkpoint via kinase inhibitors, we confirmed that this pathway is essential for organoids. Interestingly, our results suggest that specifically targeting aurora kinase A (AURKA) may represent a promising therapeutic strategy since our cells were equally sensitive to Alisertib treatment as both spheroids and organoids. Our findings emphasize the importance of studying cellular adaptations of EOC cells, as there may be different therapeutic targets depending on the step of EOC disease progression.
Keywords: G2/M checkpoint pathway; cancer dormancy; epithelial ovarian cancer; high-grade serous; organoids; spheroids; transcriptome.
Conflict of interest statement
The authors declare no conflicts of interest for this study.
Figures


References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

