Calponin 3 Regulates Myoblast Proliferation and Differentiation Through Actin Cytoskeleton Remodeling and YAP1-Mediated Signaling in Myoblasts
- PMID: 39851570
- PMCID: PMC11764405
- DOI: 10.3390/cells14020142
Calponin 3 Regulates Myoblast Proliferation and Differentiation Through Actin Cytoskeleton Remodeling and YAP1-Mediated Signaling in Myoblasts
Abstract
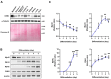
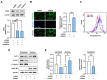
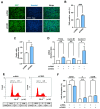
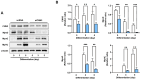
An actin-binding protein, known as Calponin 3 (CNN3), modulates the remodeling of the actin cytoskeleton, a fundamental process for the maintenance of skeletal muscle homeostasis. Although the roles of CNN3 in actin remodeling have been established, its biological significance in myoblast differentiation remains largely unknown. This study investigated the functional significance of CNN3 in myogenic differentiation, along with its effects on actin remodeling and mechanosensitive signaling in C2C12 myoblasts. CNN3 knockdown led to a marked increase in filamentous actin, which promoted the nuclear localization of Yes-associated protein 1 (YAP1), a mechanosensitive transcriptional coactivator required for response to the mechanical cues that drive cell proliferation. Subsequently, CNN3 depletion enhanced myoblast proliferation by upregulating the expression of the YAP1 target genes related to cell cycle progression, such as cyclin B1, cyclin D1, and PCNA. According to a flow cytometry analysis, CNN3-deficient cells displayed higher S and G2/M phase fractions, which concurred with elevated proliferation rates. Furthermore, CNN3 knockdown impaired myogenic differentiation, as evidenced by reduced levels of MyoD, MyoG, and MyHC, key markers of myogenic commitment and maturation, and immunocytochemistry showed that myotube formation was diminished in CNN3-suppressed cells, which was supported by lower differentiation and fusion indices. These findings reveal that CNN3 is essential for myogenic differentiation, playing a key role in regulating actin remodeling and cellular localization of YAP1 to orchestrate the proliferation and differentiation in myogenic progenitor cells. This study highlights CNN3 as a critical regulator of skeletal myogenesis and suggests its therapeutic potential as a target for muscle atrophy and related disorders.
Keywords: YAP1; actin cytoskeleton remodeling; calponin; mechanotransduction; myogenic differentiation; proliferation.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
Similar articles
-
Essential Role of Cortactin in Myogenic Differentiation: Regulating Actin Dynamics and Myocardin-Related Transcription Factor A-Serum Response Factor (MRTFA-SRF) Signaling.Int J Mol Sci. 2024 Dec 18;25(24):13564. doi: 10.3390/ijms252413564. Int J Mol Sci. 2024. PMID: 39769327 Free PMC article.
-
Twinfilin-1 is an essential regulator of myogenic differentiation through the modulation of YAP in C2C12 myoblasts.Biochem Biophys Res Commun. 2022 Apr 9;599:17-23. doi: 10.1016/j.bbrc.2022.02.021. Epub 2022 Feb 8. Biochem Biophys Res Commun. 2022. PMID: 35168059
-
Rock-dependent calponin 3 phosphorylation regulates myoblast fusion.Exp Cell Res. 2013 Mar 10;319(5):633-48. doi: 10.1016/j.yexcr.2012.12.022. Epub 2012 Dec 28. Exp Cell Res. 2013. PMID: 23276748
-
Calponin isoforms CNN1, CNN2 and CNN3: Regulators for actin cytoskeleton functions in smooth muscle and non-muscle cells.Gene. 2016 Jul 1;585(1):143-153. doi: 10.1016/j.gene.2016.02.040. Epub 2016 Mar 10. Gene. 2016. PMID: 26970176 Free PMC article. Review.
-
[Interactions of proliferation and differentiation signaling pathways in myogenesis].Postepy Hig Med Dosw (Online). 2014 May 8;68:516-26. doi: 10.5604/17322693.1101617. Postepy Hig Med Dosw (Online). 2014. PMID: 24864103 Review. Polish.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

