A Truncated Multi-Thiol Aptamer-Based SARS-CoV-2 Electrochemical Biosensor: Towards Variant-Specific Point-of-Care Detection with Optimized Fabrication
- PMID: 39852074
- PMCID: PMC11763500
- DOI: 10.3390/bios15010024
A Truncated Multi-Thiol Aptamer-Based SARS-CoV-2 Electrochemical Biosensor: Towards Variant-Specific Point-of-Care Detection with Optimized Fabrication
Abstract
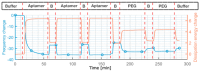
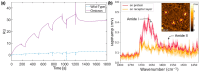
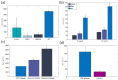
With the goal of fast and accurate diagnosis of infectious diseases, this study presents a novel electrochemical biosensor that employs a refined aptamer (C9t) for the detection of spike (S) protein SARS-CoV-2 variants in a flexible multielectrode aptasensor array with PoC capabilities. Two aptamer modifications were employed: removing the primer binding sites and including two dithiol phosphoramidite anchor molecules. Thus, reducing fabrication time from 24 to 3 h and increasing the stability and sparseness for multi-thiol aptasensors compared to a standard aptasensor using single thiols, without a reduction in aptamer density. The biosensor fabrication, optimization, and detection were verified in detail by electrochemistry, QCM-D, SPR, and XPS. The analyte-receptor binding was further confirmed spectroscopically at the level of individual molecules by AFM-IR. The aptasensor possesses a low limit of detection (8.0 fg/mL), the highest sensitivity reported for S protein (209.5 signal per concentration decade), and a wide dynamic detection range (8.0 fg/mL-38 ng/mL) in nasopharyngeal samples, covering the clinically relevant range. Furthermore, the C9t aptasensor showed high selectivity for SARS-CoV-2 S proteins over biomarkers for MERS-CoV, RSV, and Influenza. Even more, it showed a three times higher sensitivity for the Omicron in comparison to the Wuhan strain (wild type), alpha, and beta variants of the SARS-CoV-2 virus. Those results demonstrate the creation of an affordable and variant-selective refined C9t aptasensor that outperformed current rapid diagnosis tests.
Keywords: S protein; SARS-CoV-2; aptamer truncation; electrochemical aptasensor; multi-thiol.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

References
-
- Souf S. Recent Advances in diagnostic testing for viral infections. Biosci. Horiz. 2016;9:hzw010. doi: 10.1093/biohorizons/hzw010. - DOI
-
- Aylward B., Liang W., Dong X., Eckmanns T., Fisher D., Ihekweazu C., Lane C., Lee J.-K., Leung G., Lin J., et al. WHO China Joint Mission on COVID-19 Final Report. 2020. [(accessed on 21 November 2024)]. Available online: https://www.who.int/docs/default-source/coronaviruse/who-china-joint-mis....
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

