Phytophthora Species and Their Associations with Chaparral and Oak Woodland Vegetation in Southern California
- PMID: 39852452
- PMCID: PMC11766400
- DOI: 10.3390/jof11010033
Phytophthora Species and Their Associations with Chaparral and Oak Woodland Vegetation in Southern California
Abstract
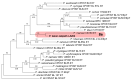
Evidence of unintended introductions of Phytophthora species into native habitats has become increasingly prevalent in California. If not managed adequately, Phytophthora species can become devastating agricultural and forest plant pathogens. Additionally, California's natural areas, characterized by a Mediterranean climate and dominated by chaparral (evergreen, drought-tolerant shrubs) and oak woodlands, lack sufficient baseline knowledge on Phytophthora biology and ecology, hindering effective management efforts. From 2018 to 2021, soil samples were collected from Angeles National Forest lands (Los Angeles County) with the objective of better understanding the diversity and distribution of Phytophthora species in Southern California. Forty sites were surveyed, and soil samples were taken from plant rhizospheres, riverbeds, and off-road vehicle tracks in chaparral and oak woodland areas. From these surveys, fourteen species of Phytophthora were detected, including P. cactorum (subclade 1a), P. multivora (subclade 2c), P. sp. cadmea (subclade 7a), P. taxon 'oakpath' (subclade 8e, first reported in this study), and several clade-6 species, including P. crassamura. Phytophthora species detected in rhizosphere soil were found underneath both symptomatic and asymptomatic plants and were most frequently associated with Salvia mellifera, Quercus agrifolia, and Salix sp. Phytophthora species were present in both chaparral and oak woodland areas and primarily in riparian areas, including detections in off-road tracks, trails, and riverbeds. Although these Mediterranean ecosystems are among the driest and most fire-prone areas in the United States, they harbor a large diversity of Phytophthora species, indicating a potential risk for disease for native Californian vegetation.
Keywords: Mediterranean vegetation; oomycete; plant pathogen.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures







Similar articles
-
Diversity of Phytophthora Species from Declining Mediterranean Maquis Vegetation, including Two New Species, Phytophthora crassamura and P. ornamentata sp. nov.PLoS One. 2015 Dec 9;10(12):e0143234. doi: 10.1371/journal.pone.0143234. eCollection 2015. PLoS One. 2015. PMID: 26649428 Free PMC article.
-
Diversity and distribution of Phytophthora species across different types of riparian vegetation in Italy with the description of Phytophthora heteromorpha sp. nov.Int J Syst Evol Microbiol. 2024 Feb;74(2). doi: 10.1099/ijsem.0.006272. Int J Syst Evol Microbiol. 2024. PMID: 38407194
-
Influence of land-cover change on the spread of an invasive forest pathogen.Ecol Appl. 2008 Jan;18(1):159-71. doi: 10.1890/07-0232.1. Ecol Appl. 2008. PMID: 18372563
-
Phytophthora ramorum: integrative research and management of an emerging pathogen in California and Oregon forests.Annu Rev Phytopathol. 2005;43:309-35. doi: 10.1146/annurev.phyto.42.040803.140418. Annu Rev Phytopathol. 2005. PMID: 16078887 Review.
-
Nitrogen critical loads and management alternatives for N-impacted ecosystems in California.J Environ Manage. 2010 Dec;91(12):2404-23. doi: 10.1016/j.jenvman.2010.07.034. Epub 2010 Aug 11. J Environ Manage. 2010. PMID: 20705383 Review.
References
Grants and funding
LinkOut - more resources
Full Text Sources

