Direct and Indirect Protein Interactions Link FUS Aggregation to Histone Post-Translational Modification Dysregulation and Growth Suppression in an ALS/FTD Yeast Model
- PMID: 39852477
- PMCID: PMC11766905
- DOI: 10.3390/jof11010058
Direct and Indirect Protein Interactions Link FUS Aggregation to Histone Post-Translational Modification Dysregulation and Growth Suppression in an ALS/FTD Yeast Model
Abstract
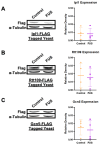
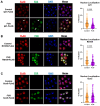
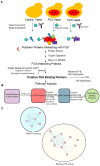
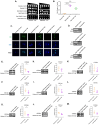
Amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD) are incurable neurodegenerative disorders sharing pathological and genetic features, including mutations in the FUS gene. FUS is an RNA-binding protein that mislocalizes to the cytoplasm and aggregates in ALS/FTD. In a yeast model, FUS proteinopathy is connected to changes in the epigenome, including reductions in the levels of H3S10ph, H3K14ac, and H3K56ac. Exploiting the same model, we reveal novel connections between FUS aggregation and epigenetic dysregulation. We show that the histone-modifying enzymes Ipl1 and Rtt109-responsible for installing H3S10ph and H3K56ac-are excluded from the nucleus in the context of FUS proteinopathy. Furthermore, we found that Ipl1 colocalizes with FUS, but does not bind it directly. We identified Nop1 and Rrp5, a histone methyltransferase and rRNA biogenesis protein, respectively, as FUS binding partners involved in the growth suppression phenotype connected to FUS proteinopathy. We propose that the nuclear exclusion of Ipl1 through indirect interaction with FUS drives the dysregulation of H3S10ph as well as H3K14ac via crosstalk. We found that the knockdown of Nop1 interferes with these processes. In a parallel mechanism, Rtt109 mislocalization results in reduced levels of H3K56ac. Our results highlight the contribution of epigenetic mechanisms to ALS/FTD and identify novel targets for possible therapeutic intervention.
Keywords: FUS; Ipl1; Nop1; Rtt109; amyotrophic lateral sclerosis; epigenetics; frontotemporal dementia; histone post-translational modifications.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

References
-
- Pang S.Y.-Y., Hsu J.S., Teo K.-C., Li Y., Kung M.H.W., Cheah K.S.E., Chan D., Cheung K.M.C., Li M., Sham P.-C., et al. Burden of Rare Variants in ALS Genes Influences Survival in Familial and Sporadic ALS. Neurobiol. Aging. 2017;58:238.e9–238.e15. doi: 10.1016/j.neurobiolaging.2017.06.007. - DOI - PubMed
-
- Debove C., Zeisser P., Salzman P.M., Powe L.K., Truffinet P. The Rilutek (Riluzole) Global Early Access Programme: An Open-Label Safety Evaluation in the Treatment of Amyotrophic Lateral Sclerosis. Amyotroph. Lateral Scler. Other Mot. Neuron Disord. 2001;2:153–158. doi: 10.1080/146608201753275508. - DOI - PubMed
-
- Brooks B.R., Jorgenson J.A., Newhouse B.J., Shefner J.M., Agnese W. Edaravone in the Treatment of Amyotrophic Lateral Sclerosis: Efficacy and Access to Therapy—A Roundtable Discussion. Am. J. Manag. Care. 2018;24:S175–S186. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

