The Development of a One-Step PCR Assay for Rapid Detection of an Attenuated Vaccine Strain of Duck Hepatitis Virus Type 3 in Korea
- PMID: 39852883
- PMCID: PMC11768531
- DOI: 10.3390/vetsci12010008
The Development of a One-Step PCR Assay for Rapid Detection of an Attenuated Vaccine Strain of Duck Hepatitis Virus Type 3 in Korea
Abstract
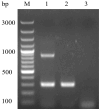
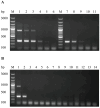
Duck hepatitis A virus type 3 (DHAV-3) is a viral pathogen that causes acute, high-mortality hepatitis in ducklings, and vaccination with attenuated live vaccines is currently the main preventive measure against it. However, differentiating infected from vaccinated animals (DIVA) is crucial for clinical diagnosis and effective disease control. This study aimed to develop a rapid mismatch amplification mutation assay PCR (MAMA-PCR) diagnostic method to simultaneously detect and differentiate between wild-type and vaccine strains. The method was specifically designed to target the critical single-nucleotide polymorphism (SNP) site (T→C at position 1143 in the VP0 gene) unique to the Korean vaccine strain AP04203-P100. MAMA-PCR demonstrated high sensitivity and specificity, with detection limits as low as 102.4 ELD50/mL for wild strains and 100.5 ELD50/mL for vaccine strains, and showed no cross-reactivity with 11 other common duck pathogens. The clinical sample results were completely consistent with those obtained using nested PCR detection and gold-standard sequencing. In summary, we successfully developed a rapid, one-step MAMA-PCR method that is more suitable for clinical diagnosis than traditional sequencing methods.
Keywords: differentiating infected from vaccinated animals; duck hepatitis A virus type 3; mismatch amplification mutation assay; rapid diagnostic.
Conflict of interest statement
Authors Hyung-Kwan Jang and Min Kang were employed by Bio Disease Control (BIOD) Co., Ltd. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as potential conflicts of interest.
Figures



References
-
- WOAH . Manual of Diagnostic Tests and Vaccines for Terrestrial Animals. 12th ed. WOAH; Paris, France: 2024. [(accessed on 4 November 2024)]. Duck Virus Hepatitis. Chapter 3.3.6. Available online: https://www.woah.org/fileadmin/Home/eng/Health_standards/tahm/3.03.06_DV....
LinkOut - more resources
Full Text Sources

