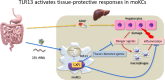
RNase T2 deficiency promotes TLR13-dependent replenishment of tissue-protective Kupffer cells
- PMID: 39853307
- PMCID: PMC11758922
- DOI: 10.1084/jem.20230647
RNase T2 deficiency promotes TLR13-dependent replenishment of tissue-protective Kupffer cells
Abstract
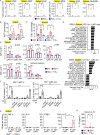
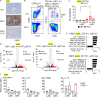
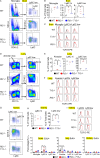
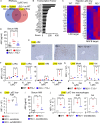
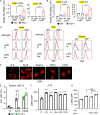
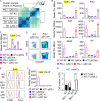
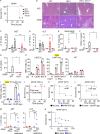
Lysosomal stress due to the accumulation of nucleic acids (NAs) activates endosomal TLRs in macrophages. Here, we show that lysosomal RNA stress, caused by the lack of RNase T2, induces macrophage accumulation in multiple organs such as the spleen and liver through TLR13 activation by microbiota-derived ribosomal RNAs. TLR13 triggered emergency myelopoiesis, increasing the number of myeloid progenitors in the bone marrow and spleen. Splenic macrophages continued to proliferate and mature into macrophages expressing the anti-inflammatory cytokine IL-10. In the liver, TLR13 activated monocytes/macrophages to proliferate and mature into monocyte-derived KCs (moKCs), in which, the liver X receptor (LXR) was activated. In accumulated moKCs, tissue clearance genes such as MerTK, AXL, and apoptosis inhibitor of macrophage (AIM) were highly expressed, while TLR-dependent production of proinflammatory cytokines was impaired. Consequently, Rnaset2-/- mice were resistant to acute liver injuries elicited by acetaminophen (APAP) and LPS with D-galactosamine. These findings suggest that TLR13 activated by lysosomal RNA stress promotes the replenishment of tissue-protective Kupffer cells.
© 2025 Sato et al.
Conflict of interest statement
Disclosures: R. Fukui reported grants from Daiichi Sankyo outside the submitted work. No other disclosures were reported.
Figures





References
-
- Akashi-Takamura, S., Furuta T., Takahashi K., Tanimura N., Kusumoto Y., Kobayashi T., Saitoh S., Adachi Y., Doi T., and Miyake K.. 2006. Agonistic antibody to TLR4/MD-2 protects mice from acute lethal hepatitis induced by TNF-alpha. J. Immunol. 176:4244–4251. 10.4049/jimmunol.176.7.4244 - DOI - PubMed
-
- Allen, R.M., Michell D.L., Cavnar A.B., Zhu W., Makhijani N., Contreras D.M., Raby C.A., Semler E.M., DeJulius C., Castleberry M., et al. . 2022. LDL delivery of microbial small RNAs drives atherosclerosis through macrophage TLR8. Nat. Cell Biol. 24:1701–1713. 10.1038/s41556-022-01030-7 - DOI - PMC - PubMed
-
- Allen, R.M., Zhao S., Ramirez Solano M.A., Zhu W., Michell D.L., Wang Y., Shyr Y., Sethupathy P., Linton M.F., Graf G.A., et al. . 2018. Bioinformatic analysis of endogenous and exogenous small RNAs on lipoproteins. J. Extracell. Vesicles. 7:1506198. 10.1080/20013078.2018.1506198 - DOI - PMC - PubMed
-
- Arai, S., Kitada K., Yamazaki T., Takai R., Zhang X., Tsugawa Y., Sugisawa R., Matsumoto A., Mori M., Yoshihara Y., et al. . 2016. Apoptosis inhibitor of macrophage protein enhances intraluminal debris clearance and ameliorates acute kidney injury in mice. Nat. Med. 22:183–193. 10.1038/nm.4012 - DOI - PubMed
-
- Bonnardel, J., T’Jonck W., Gaublomme D., Browaeys R., Scott C.L., Martens L., Vanneste B., De Prijck S., Nedospasov S.A., Kremer A., et al. . 2019. Stellate cells, hepatocytes, and endothelial cells imprint the kupffer cell identity on monocytes colonizing the liver macrophage niche. Immunity. 51:638–654.e9. 10.1016/j.immuni.2019.08.017 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

