Turning Waste into Treasure: Functionalized Biomass-Derived Carbon Dots for Superselective Visualization and Eradication of Gram-Positive Bacteria
- PMID: 39853875
- PMCID: PMC11923988
- DOI: 10.1002/advs.202411084
Turning Waste into Treasure: Functionalized Biomass-Derived Carbon Dots for Superselective Visualization and Eradication of Gram-Positive Bacteria
Abstract
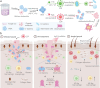
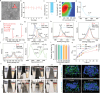
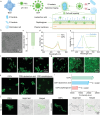
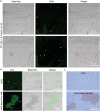
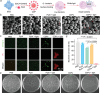
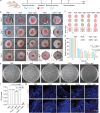
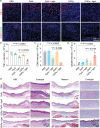
Gram-positive bacteria pose significant threats to human health, necessitating the development of targeted bacterial detection and eradication strategies. Nevertheless, current approaches often suffer from poor targeting specificity. Herein, the study utilizes purple rice lixivium to synthesize biomass carbon dots (termed BCDs) with wheat germ agglutinin-like residues for precisely targeting Gram-positive bacteria. Subsequently, fluorescein isothiocyanate (FITC) molecules are grafted onto BCDs to yield FITC-labeled BCDs (termed CDFs), which can selectively and rapidly (≤5 min) stain bacterial cell wall and particularly target the peptidoglycan component. Strikingly, CDFs achieve superselective visualization of Gram-positive bacteria even in the presence of mammalian cells and Gram-negative bacteria. Furthermore, protoporphyrin (PpIX) molecules are conjugated onto BCDs to yield PpIX-modified BCDs (termed CDPs), which can induce bacterial aggregation and in situ generate singlet oxygen for realizing enhanced antibacterial photodynamic therapy (PDT). At the minimum bactericidal concentration of CDPs (PpIX: 5 µg mL-1), CDP-mediated PDT disrupts bacterial structure and metabolism pathways, thereby affecting bacterial interactions to eradicate biofilms. Importantly, CDP-mediated PDT efficiently modulates antiinflammatory responses to promote wound healing in the bacteria-infected mice. This study underscores the significance of harnessing renewable and cost-effective biomass resources for preparing Gram-positive bacteria-targeting theranostic agents, which may find potential clinical applications in the future.
Keywords: antibacterial therapy; bacterial imaging; biomass carbon dots; cell wall‐targeting probes; wound healing.
© 2025 The Author(s). Advanced Science published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Kwon H. Y., Liu X., Choi E. G., Lee J. Y., Choi S. Y., Kim J. Y., Wang L., Park S. J., Kim B., Lee Y. A., Kim J. J., Kang N. Y., Chang Y. T., Angew. Chem. Int. Ed. 2019, 58, 8426. - PubMed
-
- a) Li X., Bai H., Yang Y., Yoon J., Wang S., Zhang X., Adv. Mater. 2019, 31, 1805092; - PubMed
- b) Zhao E., Chen Y., Chen S., Deng H., Gui C., Leung C. W. T., Hong Y., Lam J. W. Y., Tang B. Z., Adv. Mater. 2015, 27, 4931; - PubMed
- c) van Oosten M., Schäfer T., Gazendam J. A. C., Ohlsen K., Tsompanidou E., de Goffau M. C., Harmsen H. J. M., Crane L. M. A., Lim E., Francis K. P., Cheung L., Olive M., Ntziachristos V., van Dijl J. M., van Dam G. M., Nat. Commun. 2013, 4, 2584. - PubMed
-
- Li J., Lin S., Wang J., Jia S., Yang M., Hao Z., Zhang X., Chen P. R., J. Am. Chem. Soc. 2013, 135, 7330. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
