Preliminary screening of new biomarkers for sepsis using bioinformatics and experimental validation
- PMID: 39854580
- PMCID: PMC11759385
- DOI: 10.1371/journal.pone.0317608
Preliminary screening of new biomarkers for sepsis using bioinformatics and experimental validation
Abstract
Background: The morbidity and mortality of sepsis remain high, and so far specific diagnostic and therapeutic means are lacking.
Objective: To screen novel biomarkers for sepsis.
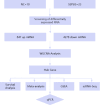
Methods: Raw sepsis data were downloaded from the Chinese National Genebank (CNGBdb) and screened for differentially expressed RNAs. Key genes with predictive value were identified through weighted correlation network analysis (WGCNA) and meta-analysis and survival analysis using multiple public databases. Core genes were analyzed for functional enrichment using Gene Set Enrichment Analysis(GSEA). The core genes were localized using single-cell sequencing. qPCR was used to validate the core genes.
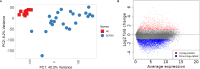
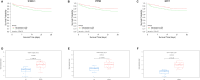
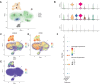
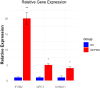
Results: Differential analysis yielded a total of 5125 mRNA. WGCNA identified 5 modules and screened 3 core genes (S100A11, QPCT, and IFITM2). The prognosis of sepsis patients was strongly linked to S100A11, QPCT, and IFITM2 based on meta-analysis and survival analysis(P < 0.05).GSEA analysis showed that S100A11, QPCT, and IFITM2 were significantly enriched in ribosome-related pathways. S100A11 and QPCT were widely distributed in all immune cells, and QPCT was mainly localized in the macrophage cell lineage. In the sepsis group, the qPCR results showed that S100A11, QPCT, and IFITM2 expression levels were significantly higher in the sepsis group(P < 0.05).
Conclusion: In this study, S100A11, QPCT, and IFITM2 were screened as new potential biomarkers for sepsis. Validated by bioinformatics analysis and qPCR, these genes are closely associated with the prognosis of sepsis patients and have potential as diagnostic and therapeutic targets.
Copyright: © 2025 Wang et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



Similar articles
-
LINC02363: a potential biomarker for early diagnosis and treatment of sepsis.BMC Immunol. 2025 Mar 15;26(1):23. doi: 10.1186/s12865-025-00702-x. BMC Immunol. 2025. PMID: 40089725 Free PMC article.
-
Exploring the prognostic and diagnostic value of lactylation-related genes in sepsis.Sci Rep. 2024 Oct 4;14(1):23130. doi: 10.1038/s41598-024-74040-0. Sci Rep. 2024. PMID: 39367086 Free PMC article.
-
Identification of key genes as potential diagnostic biomarkers in sepsis by bioinformatics analysis.PeerJ. 2024 Jun 18;12:e17542. doi: 10.7717/peerj.17542. eCollection 2024. PeerJ. 2024. PMID: 38912048 Free PMC article.
-
Molecular heterogeneity in pediatric sepsis: identification of oxidative stress-related subtypes and diagnostic biomarkers through integrated bioinformatics analysis.Toxicol Mech Methods. 2025 Jun;35(5):551-563. doi: 10.1080/15376516.2025.2466577. Epub 2025 Feb 28. Toxicol Mech Methods. 2025. PMID: 39950836
-
Bioinformatics and Machine Learning Methods Identified MGST1 and QPCT as Novel Biomarkers for Severe Acute Pancreatitis.Mol Biotechnol. 2024 May;66(5):1246-1265. doi: 10.1007/s12033-023-01026-0. Epub 2024 Jan 18. Mol Biotechnol. 2024. PMID: 38236462
References
-
- Oami T, Imaeda T, Nakada T-A, Abe T, Takahashi N, Yamao Y, et al.. Mortality analysis among sepsis patients in and out of intensive care units using the Japanese nationwide medical claims database: a study by the Japan sepsis alliance study group. J Intensive Care. 2023;11(1):2. doi: 10.1186/s40560-023-00650-x - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

