Evolution of SARS-CoV-2 in white-tailed deer in Pennsylvania 2021-2024
- PMID: 39854608
- PMCID: PMC11781694
- DOI: 10.1371/journal.ppat.1012883
Evolution of SARS-CoV-2 in white-tailed deer in Pennsylvania 2021-2024
Abstract
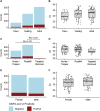
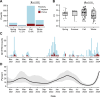
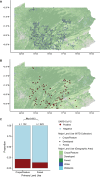
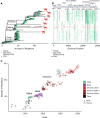
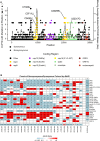
SARS-CoV-2 continues to transmit and evolve in humans and animals. White-tailed deer (Odocoileus virginianus) have been previously identified as a zoonotic reservoir for SARS-CoV-2 with high rates of infection and probable spillback into humans. Here we report sampling 1,127 white-tailed deer (WTD) in Pennsylvania, and a genomic analysis of viral dynamics spanning 1,017 days between April 2021 and January 2024. To assess viral load and genotypes, RNA was isolated from retropharyngeal lymph nodes and analyzed using RT-qPCR and viral whole genome sequencing. Samples showed a 14.64% positivity rate by RT-qPCR. Analysis showed no association of SARS-CoV-2 prevalence with age, sex, or diagnosis with Chronic Wasting Disease. From the 165 SARS-CoV-2 positive WTD, we recovered 25 whole genome sequences and an additional 17 spike-targeted amplicon sequences. The viral variants identified included 17 Alpha, 11 Delta, and 14 Omicron. Alpha largely stopped circulating in humans around September 2021, but persisted in WTD as recently as March of 2023. Phylodynamic analysis of pooled genomic data from Pennsylvania documents at least 12 SARS-CoV-2 spillovers from humans into WTD, including a recent series of Omicron spillovers. Prevalence was higher in WTD in regions with crop coverage rather than forest, suggesting an association with proximity to humans. Analysis of seasonality showed increased prevalence in winter and spring. Multiple examples of recurrent mutations were identified associated with transmissions, suggesting WTD-specific evolutionary pressures. These data document ongoing infections in white-tailed deer, probable onward transmission in deer, and a remarkable rate of new spillovers from humans.
Copyright: © 2025 Marques et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
-
- Organization WH. COVID-19 cases | WHO COVID-19 dashboard. datadot; 2024.
-
- Pereira AHB, Pereira GO, Borges JC, de Barros Silva VL, Pereira BHM, Morgado TO, et al.. A novel host of an emerging disease: SARS-CoV-2 infection in a giant anteater (Myrmecophaga tridactyla) kept under clinical care in Brazil. Ecohealth. 2022;19(4):458–62. doi: 10.1007/s10393-022-01623-6 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

