Single-cell analysis of neoplastic plasma cells identifies myeloma pathobiology mediators and potential targets
- PMID: 39855192
- PMCID: PMC11866523
- DOI: 10.1016/j.xcrm.2024.101925
Single-cell analysis of neoplastic plasma cells identifies myeloma pathobiology mediators and potential targets
Abstract
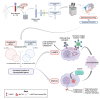
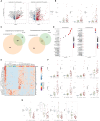
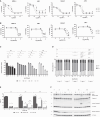
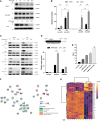
Multiple myeloma is a clonal plasma cell (PC) dyscrasia that arises from precursors and has been studied utilizing approaches focused on CD138+ cells. By combining single-cell RNA sequencing (scRNA-seq) with scB-cell receptor sequencing (scBCR-seq), we differentiate monoclonal/neoplastic from polyclonal/normal PCs and find more dysregulated genes, especially in precursor patients, than we would have by analyzing bulk PCs. To determine whether this approach can identify oncogenes that contribute to disease pathobiology, mitotic arrest deficient-2 like-1 (MAD2L1) and S-adenosylmethionine synthase isoform type-2 (MAT2A) are validated as targets with drug-like molecules that suppress myeloma growth in preclinical models. Moreover, functional studies show a role of lysosomal-associated membrane protein family member-5 (LAMP5), which is uniquely expressed in neoplastic PCs, in tumor progression and aggressiveness via interactions with c-MYC. Finally, a monoclonal antibody recognizing cell-surface LAMP5 shows efficacy as an antibody-drug conjugate and in a chimeric antigen receptor-guided T-cell format. These studies provide additional insights into myeloma biology and identify potential targeted therapeutic approaches that can be applied to reverse myeloma progression.
Keywords: LAMP5; MAT2A; disease progression; myeloma precursors; scBCR-seq; scRNA-seq.
Copyright © 2025 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests H.C.L. has provided consultancy services to AbbVie, Bristol Myers Squibb, Genentech, Janssen, Regeneron, GlaxoSmithKline, Sanofi, Takeda Pharmaceuticals, and Allogene Therapeutics and has received research funding from Amgen, Bristol Myers Squibb, Janssen, GlaxoSmithKline, Regeneron, and Takeda Pharmaceuticals. K.K.P. declares research support from Celgene, a wholly owned subsidiary of Bristol Myers Squibb. R.Z.O. declares research funding unrelated to this work from Heidelberg Pharma AG, Asylia Therapeutics, and Biotheryx. Also, R.Z.O. has served on advisory boards for Amgen, Inc., Bristol Myers Squibb, Celgene, EcoR1 Capital LLC, Forma Therapeutics, Genzyme, GSK Biologicals, Ionis Pharmaceuticals, Inc., Janssen Biotech, Juno Therapeutics, Kite Pharma, Legend Biotech USA, Molecular Partners, Sanofi-Aventis, Servier, and Takeda Pharmaceuticals North America, Inc. and is a founder of Asylia Therapeutics, Inc., with an equity interest. R.Z.O., D.E.S., H.W., and L.Y.M.R. declare a provisional patent application around the anti-LAMP5 antibody discussed herein.
Figures



References
-
- Institute N.C. 2024. Cancer Stat Facts: Myeloma. https://seer.cancer.gov/statfacts/html/mulmy.html.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

