Natural repeated backcrosses lead to triploidy and tetraploidy in parthenogenetic butterfly lizards (Leiolepis: Agamidae)
- PMID: 39856096
- PMCID: PMC11760361
- DOI: 10.1038/s41598-024-83300-y
Natural repeated backcrosses lead to triploidy and tetraploidy in parthenogenetic butterfly lizards (Leiolepis: Agamidae)
Abstract
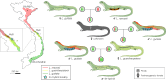
Obligatory parthenogenesis in vertebrates is restricted to squamate reptiles and evolved through hybridisation. Parthenogens can hybridise with sexual species, resulting in individuals with increased ploidy levels. We describe two successive hybridisations of the parthenogenetic butterfly lizards (genus Leiolepis) in Vietnam with a parental sexual species. Contrary to previous proposals, we document that parthenogenetic L. guentherpetersi has mitochondrial DNA and two haploid sets from L. guttata and one from L. reevesii, suggesting that it is the result of a backcross of a parthenogenetic L. guttata × L. reevesii hybrid with a L. guttata male increasing ploidy from 2n to 3n. Within the range of L. guentherpetersi, we found an adult tetraploid male with three L. guttata and one L. reevesii haploid genomes. It probably originated from fertilisation of an unreduced triploid L. guentherpetersi egg by a L. guttata sperm. Although its external morphology resembles that of the maternal species, it possessed exceptionally large erythrocytes and was likely sterile. As increased ploidy level above triploidy or tetraploidy appears to be harmful for amniotes, all-female asexual lineages should evolve a strategy to prevent incorporation of other haploid genomes from a sexual species by avoiding fertilisation by sexual males.
Keywords: Leiolepis; Hybridisation; Meiosis; Parthenogenesis; Tetraploidy; Vietnam.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests. Ethical approval: The study was conducted according to the guidelines of the Declaration of Helsinki and approved by the Institutional Review Board (or Ethics Committee) of the Zoological Institute Russian Academy of Sciences (protocol No.1-3-15-06-2021, 15 June 2021); by the Experimental Animal Ethics Committee of A.N. Severtsov Institution of Ecology and Evolution, N48 27 May, 2021 and by Vietnam Academy of Science and Technology – Institute of Genome research. (protocol No.115/QĐ-NCHG, 4th July 2021, based on the Vietnam Museum of Nature - Academy of Sciences and Vietnam Technology i (No. 639/BTTNVN dated September 12, 2019). All applicable international, national and institutional guidelines for the care and use of animals were followed during this research.
Figures



References
-
- Kearney, M., Fujita, M. K. & Ridenour, J. Lost sex in reptiles: Constraints and correlations. In Lost Sex: The Evolutionary Biology of Parthenogenesis (eds Schön, I. et al.) 447–474 (Springer Scientific, 2009). 10.1007/978-90-481-2770-2_21.
-
- Shimizu, Y., Shibata, N., Sakaizumi, M. & Yamashita, M. Production of diploid eggs through premeiotic endomitosis in the hybrid medaka between Oryzias latipes and O. curvinotus. Zool. Sci.17, 951–958. 10.2108/zsj.17.951 (2000). - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

