Multiomics approach identifies dysregulated lipidomic and proteomic networks in Parkinson's disease patients mutated in TMEM175
- PMID: 39856101
- PMCID: PMC11760379
- DOI: 10.1038/s41531-024-00853-5
Multiomics approach identifies dysregulated lipidomic and proteomic networks in Parkinson's disease patients mutated in TMEM175
Abstract
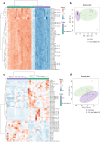
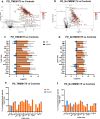
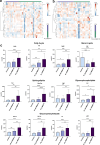
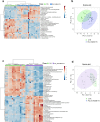
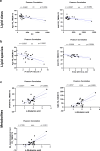
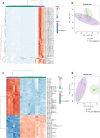
Parkinson's disease (PD) represents one of the most frequent neurodegenerative disorders for which clinically useful biomarkers remain to be identified and validated. Here, we adopted an untargeted omics approach to disclose lipidomic, metabolomic and proteomic alterations in plasma and in dermal fibroblasts of PD patients carrying mutations in TMEM175 gene. We revealed a wide dysregulation of lysosome, autophagy, and mitochondrial pathways in these patients, supporting a role of this channel in regulating these cellular processes. The most significant altered lipid classes were Fatty acyls, Glycerophospholipids and Phosphosphingolipids. The plasma level of Phosphatidylcholines (PC) and Phosphatidylinositol (PI) 34:1 significantly correlated with an earlier age at onset of the disease in TMEM175 patients (p = 0.008; p = 0.006). In plasma we also observed altered amino acids metabolic pathways in PD patients. We highlighted that increased level of L-glutamate strongly correlated (p < 0.001) with the severity of motor and non-motor symptoms in PD_TMEM175 patients. In dermal fibroblasts, we disclosed alterations of proteins involved in lipids biosynthesis (PAG15, PP4P1, GALC, FYV1, PIGO, PGPS1, PLPP1), in the insulin pathway (IGF2R), in mitochondrial metabolism (ACD10, ACD11, ACADS) and autophagy (RAB7L). Interestingly, we quantified 43 lysosomal or lysosomal-related proteins, which were differentially modulated between TMEM175 patients and controls. Integrative correlation analysis of proteome and lipidome of PD_TMEM175 cellular models identified a strong positive correlation of 13 proteins involved in biosynthetic processes with PC and Ceramides. Altogether, these data provide novel insights into the molecular and metabolic alterations underlying TMEM175 mutations and may be relevant for PD prediction, diagnosis and treatment.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures







References
-
- Bloem, B. R., Okun, M. S. & Klein, C. Parkinson’s disease. Lancet397, 2284–2303 (2021). - PubMed
-
- Braak, H. et al. Staging of brain pathology related to sporadic Parkinson’s disease. Neurobiol. Aging24, 197–211 (2003). - PubMed
-
- Schapira, A. H. V., Chaudhuri, K. R. & Jenner, P. Erratum: Non-motor features of Parkinson disease. Nat. Rev. Neurosci.18, 509–509 (2017). - PubMed
Grants and funding
- PNRR-MAD-2022-12375960/Ministero della Salute (Ministry of Health, Italy)
- RF 2019-12370224/Ministero della Salute (Ministry of Health, Italy)
- Ricerca corrente/Ministero della Salute (Ministry of Health, Italy)
- PNRR-MAD-2022-12375960 Next Generation EU/Ministero della Salute (Ministry of Health, Italy)
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

