Integrative proteogenomic analysis identifies COL6A3-derived endotrophin as a mediator of the effect of obesity on coronary artery disease
- PMID: 39856218
- PMCID: PMC11821532
- DOI: 10.1038/s41588-024-02052-7
Integrative proteogenomic analysis identifies COL6A3-derived endotrophin as a mediator of the effect of obesity on coronary artery disease
Abstract
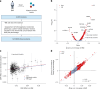
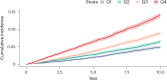
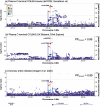
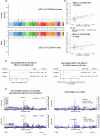
Obesity strongly increases the risk of cardiometabolic diseases, yet the underlying mediators of this relationship are not fully understood. Given that obesity strongly influences circulating protein levels, we investigated proteins mediating the effects of obesity on coronary artery disease, stroke and type 2 diabetes. By integrating two-step proteome-wide Mendelian randomization, colocalization, epigenomics and single-cell RNA sequencing, we identified five mediators and prioritized collagen type VI α3 (COL6A3). COL6A3 levels were strongly increased by body mass index and increased coronary artery disease risk. Notably, the carboxyl terminus product of COL6A3, endotrophin, drove this effect. COL6A3 was highly expressed in disease-relevant cell types and tissues. Finally, we found that body fat reduction could reduce plasma levels of COL6A3-derived endotrophin, indicating a tractable way to modify endotrophin levels. In summary, we provide actionable insights into how circulating proteins mediate the effects of obesity on cardiometabolic diseases and prioritize endotrophin as a potential therapeutic target.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: J.B.R. has served as an advisor to GlaxoSmithKline and Deerfield Capital. J.B.R.’s institution has received investigator-initiated grant funding from Eli Lilly, GlaxoSmithKline, and Biogen for projects unrelated to this research. J.B.R. is the CEO of 5 Prime Sciences ( www.5primesciences.com ), which provides research services for biotech, pharma and venture capital companies for projects unrelated to this research. T.L., Y.C. and V.F. are employees of 5 Prime Sciences. The remaining authors declare no competing interests.
Figures








References
MeSH terms
Substances
Grants and funding
- 169303/Gouvernement du Canada | Instituts de Recherche en Santé du Canada | CIHR Skin Research Training Centre (Skin Research Training Centre)
- 365825/Gouvernement du Canada | Instituts de Recherche en Santé du Canada | CIHR Skin Research Training Centre (Skin Research Training Centre)
- K99 HL169733/HL/NHLBI NIH HHS/United States
- 100558/Gouvernement du Canada | Instituts de Recherche en Santé du Canada | CIHR Skin Research Training Centre (Skin Research Training Centre)
- 409511/Gouvernement du Canada | Instituts de Recherche en Santé du Canada | CIHR Skin Research Training Centre (Skin Research Training Centre)
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

