A network of interacting ciliary tip proteins with opposing activities imparts slow and processive microtubule growth
- PMID: 39856351
- PMCID: PMC12170345
- DOI: 10.1038/s41594-025-01483-y
A network of interacting ciliary tip proteins with opposing activities imparts slow and processive microtubule growth
Abstract
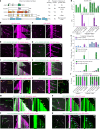
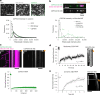
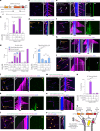
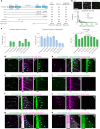
Cilia are motile or sensory organelles present on many eukaryotic cells. Their formation and function rely on axonemal microtubules, which exhibit very slow dynamics, but the underlying mechanisms are largely unexplored. Here we reconstituted in vitro the individual and collective activities of the ciliary tip module proteins CEP104, CSPP1, TOGARAM1, ARMC9 and CCDC66, which interact with each other and with microtubules and, when mutated in humans, cause ciliopathies such as Joubert syndrome. We show that CEP104, a protein with a tubulin-binding TOG domain, and its luminal partner CSPP1 inhibit microtubule growth and shortening. Another TOG-domain protein, TOGARAM1, overcomes growth inhibition imposed by CEP104 and CSPP1. CCDC66 and ARMC9 do not affect microtubule dynamics but act as scaffolds for their partners. Cryo-electron tomography demonstrated that, together, ciliary tip module members form plus-end-specific cork-like structures that reduce protofilament flaring. The combined effect of these proteins is very slow processive microtubule elongation, which recapitulates axonemal dynamics in cells.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures










References
-
- Klena, N. & Pigino, G. Structural biology of cilia and intraflagellar transport. Annu. Rev. Cell Dev. Biol.38, 103–123 (2022). - PubMed
-
- Badano, J. L., Mitsuma, N., Beales, P. L. & Katsanis, N. The ciliopathies: an emerging class of human genetic disorders. Annu. Rev. Genomics Hum. Genet.7, 125–148 (2006). - PubMed
-
- Deretic, J., Odabasi, E. & Firat-Karalar, E. N. The multifaceted roles of microtubule-associated proteins in the primary cilium and ciliopathies. J. Cell Sci.136, jcs261148 (2023). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

