Rapid detection of carbapenem-resistant Escherichia coli and carbapenem-resistant Klebsiella pneumoniae in positive blood cultures via MALDI-TOF MS and tree-based machine learning models
- PMID: 39856543
- PMCID: PMC11760114
- DOI: 10.1186/s12866-025-03755-5
Rapid detection of carbapenem-resistant Escherichia coli and carbapenem-resistant Klebsiella pneumoniae in positive blood cultures via MALDI-TOF MS and tree-based machine learning models
Abstract
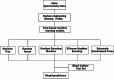
Background: Bloodstream infection (BSI) is a systemic infection that predisposes individuals to sepsis and multiple organ dysfunction syndrome. Early identification of infectious agents and determination of drug-resistant phenotypes can help patients with BSI receive timely, effective, and targeted treatment and improve their survival. This study was based on matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS), Decision Tree (DT), Random Forest (RF), Gradient Boosting Machine (GBM), eXtreme Gradient Boosting (XGBoost), and Extremely Randomized Trees (ERT) models were constructed to classify carbapenem-resistant Escherichia coli (CREC) and carbapenem-resistant Klebsiella pneumoniae (CRKP). Bacterial species were identified by MALDI-TOF MS in positive blood cultures isolated via the serum isolation gel method, and E. coli and K. pneumoniae in positive blood cultures were collected and placed into machine learning models to predict susceptibility to carbapenems. The aim of this study was to provide rapid detection of CREC and CRKP in blood cultures, to shorten the turnaround time for laboratory reporting, and to provide a basis for early clinical intervention and rational use of antibiotics.
Results: The collected MALDI-TOF MS data of 640 E. coli and 444 K. pneumoniae were analysed by machine learning algorithms. The area under the receiver operating characteristic curve (AUROC) for the diagnosis of E. coli susceptibility to carbapenems by the DT, RF, GBM, XGBoost, and ERT models were 0.95, 1.00, 0.99, 0.99, and 1.00, respectively, and the accuracy in predicting 149 E. coli-positive blood cultures were 0.89, 0.92, 0.90, 0.92, and 0.86, respectively. The AUROC for the diagnosis of K. pneumoniae susceptibility to carbapenems by the DT, RF, GBM, XGBoost, and ERT models were 0.78, 0.95, 0.93, 0.90, and 0.95, respectively, and the accuracy in predicting 127 K. pneumoniae-positive blood cultures were 0.76, 0.86, 0.81, 0.80, and 0.76, respectively.
Conclusions: Machine learning models constructed by MALDI-TOF MS were able to directly predict the susceptibility of E. coli and K. pneumoniae in positive blood cultures to carbapenems. This rapid identification of CREC and CRKP reduces detection time and contributes to early warning and response to potential antibiotic resistance problems in the clinic.
Clinical trial number: Not applicable.
Keywords: Escherichia coli; Klebsiella pneumoniae; Blood cultures; MALDI-TOF MS; Machine learning.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethical approval: Not applicable. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures



Similar articles
-
Prediction of methicillin-resistant Staphylococcus aureus and carbapenem-resistant Klebsiella pneumoniae from flagged blood cultures by combining rapid Sepsityper MALDI-TOF mass spectrometry with machine learning.Int J Antimicrob Agents. 2023 Dec;62(6):106994. doi: 10.1016/j.ijantimicag.2023.106994. Epub 2023 Oct 4. Int J Antimicrob Agents. 2023. PMID: 37802231
-
Rapid Identification and Typing of Carbapenem-Resistant Klebsiella pneumoniae Using MALDI-TOF MS and Machine Learning.Microb Biotechnol. 2025 Jun;18(6):e70184. doi: 10.1111/1751-7915.70184. Microb Biotechnol. 2025. PMID: 40522100 Free PMC article.
-
Rapid identification of carbapenem-resistant Klebsiella pneumoniae based on matrix-assisted laser desorption ionization time-of-flight mass spectrometry and an artificial neural network model.J Biomed Sci. 2023 Apr 17;30(1):25. doi: 10.1186/s12929-023-00918-2. J Biomed Sci. 2023. PMID: 37069555 Free PMC article.
-
Direct prediction of carbapenem-resistant, carbapenemase-producing, and colistin-resistant Klebsiella pneumoniae isolates from routine MALDI-TOF mass spectra using machine learning and outcome evaluation.Int J Antimicrob Agents. 2023 Jun;61(6):106799. doi: 10.1016/j.ijantimicag.2023.106799. Epub 2023 Mar 31. Int J Antimicrob Agents. 2023. PMID: 37004755
-
Systematic review of carbapenem-resistant Enterobacteriaceae causing neonatal sepsis in China.Ann Clin Microbiol Antimicrob. 2019 Nov 14;18(1):36. doi: 10.1186/s12941-019-0334-9. Ann Clin Microbiol Antimicrob. 2019. PMID: 31727088 Free PMC article.
Cited by
-
Application of metagenomic next-generation sequencing in treatment guidance for deep neck space abscess.BMC Microbiol. 2025 Mar 26;25(1):166. doi: 10.1186/s12866-025-03890-z. BMC Microbiol. 2025. PMID: 40133828 Free PMC article.
References
-
- Goto M, Al-Hasan MN. Overall burden of bloodstream infection and nosocomial bloodstream infection in North America and Europe[J]. Clin Microbiol Infect. 2013;19(6):501–9. 10.1111/1469-0691.12195. - PubMed
-
- Kern WV, Rieg S. Burden of bacterial bloodstream infection-a brief update on epidemiology and significance of multidrug-resistant pathogens[J]. Clin Microbiol Infect. 2020;26(2):151–7. 10.1016/j.cmi.2019.10.031. - PubMed
-
- Lawandi A. Bloodstream infections in Sepsis: better the Devil you Know[J]. Crit Care Med. 2023;51(9):1261–3. 10.1097/CCM.0000000000005918. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

