Study on Fermentation Preparation, Stability, and Angiotensin-Converting Enzyme Inhibitory Activity of Tomato Pomace Peptide
- PMID: 39856812
- PMCID: PMC11765387
- DOI: 10.3390/foods14020145
Study on Fermentation Preparation, Stability, and Angiotensin-Converting Enzyme Inhibitory Activity of Tomato Pomace Peptide
Abstract
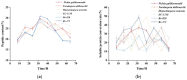
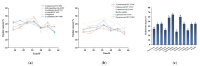
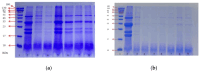
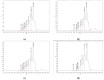
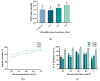
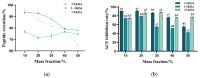
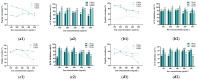
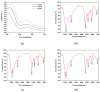
The substantial quantity of discarded tomato pomace (TP) results in the waste of valuable resources. This study utilizes these tomato by-products by mixing them with water in a specific proportion and fermenting the mixture in two stages: first with yeast, and then with lactic acid bacteria. The most suitable microbial strains for TP fermentation were identified by evaluating parameters such as peptide content, degree of hydrolysis, and gel electrophoresis analysis. Subsequently, tomato pomace peptides (TPPs) were separated into peptides of different molecular weights using ultrafiltration. The IC50 values, ACE inhibitory activities, and in vitro stability of these peptides were compared, and their secondary structures and microstructures were characterized. The results indicated that the soluble protein concentration increased from 26.25 mg/g to 39.03 mg/g after 32 h of fermentation with strain RV171. After an additional 32 h of fermentation with Bifidobacterium thermophilum, the peptide content reached 49.18 ± 0.43%. SDS-PAGE gel electrophoresis showed that the TPP molecular weights were predominantly below 10 kDa. The IC50 results demonstrated that fractions with smaller molecular weights exhibited greater ACE inhibitory activities. Structural analysis confirmed that the TP hydrolysate was indeed a peptide.
Keywords: ACE inhibitory activity; peptide; stability; structure characterization; tomato pomace; two-stage fermentation.
Conflict of interest statement
Author Keping Chen was employed by the company Xinjiang Huize Food Limited Liability Company. He participated in Supervision, project administration in the study. The role of the company was that of the Chairman of the Board of Directors. The authors declare that this study received funding from Keping Chen. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
-
- Yu S., Wang W., Bu T., Zhao R., Niu R., Liu L., Li J., Wu J., Liu D. Digestion, Absorption, and Transport Properties of Soy-Fermented Douchi Hypoglycemic Peptides VY and SFLLR under Simulated Gastrointestinal Digestion and Caco-2 Cell Monolayers. Food Res. Int. 2023;164:112340. doi: 10.1016/j.foodres.2022.112340. - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

