DICER1: The Argonaute Endonuclease Family Member and Its Role in Pediatric and Youth Pathology
- PMID: 39857323
- PMCID: PMC11761906
- DOI: 10.3390/biology14010093
DICER1: The Argonaute Endonuclease Family Member and Its Role in Pediatric and Youth Pathology
Abstract
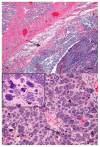
In 2001, two enzyme-encoding genes were recognized in the fruit fly Drosophila melanogaster. The genetic material, labeled Dicer-1 and Dicer-2, encodes ribonuclease-type enzymes with slightly diverse target substrates. The human orthologue is DICER1. It is a gene, which has been positioned on chromosome 14q32.13. It contains 27 exons, which are linking the two enzyme domains. DICER1 is found in all organ systems. It has been proved that it is paramount in human development. The protein determined by DICER1 is a ribonuclease (RNase). This RNase belongs to the RNase III superfamily, formally known as 'endoribonuclease'. It has been determined that the function of RNase III proteins is set to identify and degrade double-stranded molecules of RNA. DICER1 is a vital "housekeeping" gene. The multi-domain enzyme is key for small RNA processing. This enzyme functions in numerous pathways, including RNA interference paths, DNA damage renovation, and response to viruses. At the protein level, DICER is also involved in several human diseases, of which the pleuro-pulmonary blastoma is probably the most egregious entity. Numerous studies have determined the full range of DICER1 functions and the corresponding relationship to tumorigenic and non-neoplastic diseases. In fact, genetic mutations (somatic and germline) have been detected in DICER1 and are genetically associated with at least two clinical syndromes: DICER1 syndrome and GLOW syndrome. The ubiquity of this enzyme in the human body makes it an exquisite target for nanotechnology-supported therapies and repurposing drug approaches.
Keywords: DICER1; biology; molecular genetics; oncogene.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
Publication types
LinkOut - more resources
Full Text Sources

