EIF4A3-Mediated circ_0008126 Inhibits the Progression and Metastasis of Gastric Cancer by Modulating the APC/β-Catenin Pathway
- PMID: 39858034
- PMCID: PMC11763408
- DOI: 10.3390/cancers17020253
EIF4A3-Mediated circ_0008126 Inhibits the Progression and Metastasis of Gastric Cancer by Modulating the APC/β-Catenin Pathway
Abstract
Background: Mounting evidence exhibits circRNAs as critical regulators in the progression of many tumors. The regulatory function and potential mechanism by which circ_0008126 in gastric cancer (GC) is unknown.
Methods: To validate and analyze the expression levels and clinical values of circ_0008126 in GC patients, the biological phenotypes of circ_0008126 in GC were investigated in vitro and in vivo. The roles and effects of circ_0008126 on miR-502-5p, EIF4A3, and APC in GC cells were explored using rescue experiment, RNA stability assay, RNA pull-down, dual-luciferase reporter, RNA immunoprecipitation (RIP), RNA FISH, immunofluorescence (IF), and TOP/Flash and FOP/Flash assays.
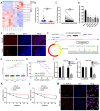
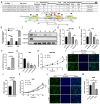
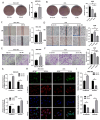
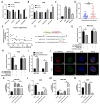
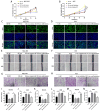
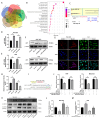
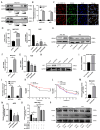
Results: Circ_0008126 expression levels were prominently down-regulated in GC tissues and cells. Importantly, low expression of circ_0008126 was relevant to the more lymphatic metastasis, advanced TNM stage, and poor survival period in patients with GC. Functionally, circ_0008126 inhibited GC cell proliferative activity, metastatic ability, and epithelial-mesenchymal transition (EMT) in vitro and vivo. Mechanistically, we verified that EIF4A3 can mediate the formation of circ_0008126, and circ_0008126 could competitively bind miR-502-5p and alleviate its role and effect on APC, thus inactivating the β-catenin pathway in GC. Additionally, circ_0008126 was determined to increase the stability of APC mRNA by interacting with cytoplasmic EIF4A3 protein and then enhancing the APC expression.
Conclusions: These data demonstrate that EIF4A3-mediated circ_0008126 could regulate the APC expression and inactivate the β-catenin pathway partly by binding to miR-502-5p and EIF4A3, thus inhibiting the tumorigenesis and development of GC.
Keywords: APC; EIF4A3; circ_0008126; gastric cancer; miR-502-5p.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Circular RNA circFGD4 suppresses gastric cancer progression via modulating miR-532-3p/APC/β-catenin signalling pathway.Clin Sci (Lond). 2020 Jul 17;134(13):1821-1839. doi: 10.1042/CS20191043. Clin Sci (Lond). 2020. PMID: 32633323
-
EIF4A3-regulated circ_0087429 can reverse EMT and inhibit the progression of cervical cancer via miR-5003-3p-dependent upregulation of OGN expression.J Exp Clin Cancer Res. 2022 May 5;41(1):165. doi: 10.1186/s13046-022-02368-4. J Exp Clin Cancer Res. 2022. PMID: 35513835 Free PMC article.
-
Hypoxia-enhanced YAP1-EIF4A3 interaction drives circ_0007386 circularization by competing with CRIM1 pre-mRNA linear splicing and promotes non-small cell lung cancer progression.J Exp Clin Cancer Res. 2024 Jul 20;43(1):200. doi: 10.1186/s13046-024-03116-6. J Exp Clin Cancer Res. 2024. PMID: 39030638 Free PMC article.
-
LINC01133 as ceRNA inhibits gastric cancer progression by sponging miR-106a-3p to regulate APC expression and the Wnt/β-catenin pathway.Mol Cancer. 2018 Aug 22;17(1):126. doi: 10.1186/s12943-018-0874-1. Mol Cancer. 2018. PMID: 30134915 Free PMC article.
-
EIF4A3-induced circ_0084615 contributes to the progression of colorectal cancer via miR-599/ONECUT2 pathway.J Exp Clin Cancer Res. 2021 Jul 12;40(1):227. doi: 10.1186/s13046-021-02029-y. J Exp Clin Cancer Res. 2021. PMID: 34253241 Free PMC article.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

