Remodeling of Mitochondria-Endoplasmic Reticulum Contact Sites Accompanies LUHMES Differentiation
- PMID: 39858520
- PMCID: PMC11764118
- DOI: 10.3390/biom15010126
Remodeling of Mitochondria-Endoplasmic Reticulum Contact Sites Accompanies LUHMES Differentiation
Abstract
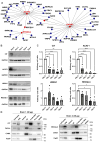
Neural progenitor cells (NPCs) are often used to study the subcellular mechanisms underlying differentiation into neurons in vitro. Works published to date have focused on the pathways that distinguish undifferentiated NPCs from mature neurons, neglecting the earlier and intermediate stages of this process. Current evidence suggests that mitochondria interaction with the ER is fundamental to a wide range of intracellular processes. However, it is not clear whether and how the mitochondria-ER interactions differ between NPCs and their differentiated counterparts. Here we take advantage of the widely used NPC line LUHMES to provide hints on the mitochondrial dynamic trait changes that occur during the first stage of their maturation into dopaminergic-like neurons. We observed that the morphology of mitochondria, their interaction with the ER, and the expression of several mitochondria-ER contact site resident proteins change, which suggests the potential contribution of mitochondria dynamics to NPC differentiation. Further studies will be needed to explore in depth these changes, and their functional outcomes, which may be relevant to the scientific community focusing on embryonic neurogenesis and developmental neurotoxicity.
Keywords: LUHMES; MERCs; Neural precursor cells; differentially expressed genes; mitochondria; mitochondria–ER contact sites.
Conflict of interest statement
Camilla Bean is a guest editor of the special issue.
Figures



References
-
- Choi H.W., Kim J.H., Chung M.K., Hong Y.J., Jang H.S., Seo B.J., Jung T.H., Kim J.S., Chung H.M., Byun S.J., et al. Mitochondrial and Metabolic Remodeling during Reprogramming and Differentiation of the Reprogrammed Cells. Stem Cells Dev. 2015;24:1366–1373. doi: 10.1089/scd.2014.0561. - DOI - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

