Comparative Analysis of Complete Chloroplast Genomes and Phylogenetic Relationships of 21 Sect. Camellia (Camellia L.) Plants
- PMID: 39858596
- PMCID: PMC11764880
- DOI: 10.3390/genes16010049
Comparative Analysis of Complete Chloroplast Genomes and Phylogenetic Relationships of 21 Sect. Camellia (Camellia L.) Plants
Abstract
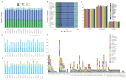
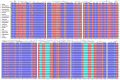
Background: Section Camellia is the most diverse group in the genus Camellia L., and this group of plants has a long history of cultivation in China as popular ornamental flowers and oil plants. Sect. Camellia plants present diverse morphological variations and complexity among species, resulting in uncertainty in the classification of species, which has resulted in a degree of inconvenience and confusion in the use of plant resources and research. Methods: Here, We sequenced and assembled the chloroplast genomes of 6 sect. Camellia and performed comparative chloroplast genome analysis and phylogenetic studies combined with 15 existing sect. Camellia plants. Results: The chloroplast genome of 21 species in sect. Camellia species were quadripartite with length of 156,587-157,068 bp base pairs (bp), and a highly conserved and moderately differentiated chloroplast genome arrangement. The 21 sect. Camellia chloroplast genomes were similar to those of angiosperms, with high consistency in gene number, gene content and gene structure. After the annotation process, we identified a total of 132 genes, specifically 87 sequences coding for proteins (CDS), 37 transfer RNA (tRNA) genes, and 8 ribosomal RNA (rRNA) genes. The ycf1 gene in 21 species of the sect. Camellia was present only in the small single-copy/inverted repeat of a (SSC/IRa) region. Sequence variation was greater in the large single-copy (LSC) region than in the IR region, and the majority of the protein-coding genes presented high codon preferences. The chloroplast genomes of 21 plant species exhibit relatively conserved SC (single copy region)/IR (inverted repeat region) boundaries. We detected a total of 2975 single sequence repeats (SSRs) as well as 833 dispersed nuclear elements (INEs). Among these SSRs, A/T repeats and AT/AT repeats dominated, while among INEs, forward repeats and palindromic repeats predominated. Codon usage frequencies were largely similar, with 30 high-frequency codons detected. Comparative analysis revealed five hotspot regions (rps16, psaJ, rpl33, rps8, and rpl16) and two gene intervals (atpH-atpI and petD-rpoA) in the cp genome, which can be used as potential molecular markers. In addition, the phylogenetic tree constructed from the chloroplast genome revealed that these 21 species and Camellia oleifera aggregated into a single branch, which was further subdivided into two evolutionarily independent sub-branches. Conclusions: It was confirmed that sect. Camellia and C. oleifera Abel are closely related in Camellia genus. These findings will enhance our knowledge of the sect. Camellia of plants, deepen our understanding of their genetic characteristics and phylogenetic pathways, and provide strong support for the scientific development and rational utilization of the plant resources of the sect. Camellia.
Keywords: chloroplast genomes; comparative genomics; phylogenetics; sect. Camellia.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures





References
-
- Chang H., Ren S. Flora of China. Science Press; Beijing, China: 1998.
-
- Chang H. Flora Reipublicae Popularis Sinicae. Science Press; Beijing, China: 1998. Camellia; pp. 37–48.
-
- Chang H. Systematic study of the genus Camellia. J. Sun Yatsen Univ. Forum. 1981:108–125.
-
- Min T., Zhong Y. A revision of genus Camellia sect. Tuberculata. Acta Bot. Yunnanica. 1993;15:123–130.
-
- Min T., Zhang W. The evolution and distribution of genus Camellia. Acta Bot. Yunnanica. 1996;18:1–13.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

