A Rewired NADPH-Dependent Redox Shuttle for Testing Peroxisomal Compartmentalization of Synthetic Metabolic Pathways in Komagataella phaffii
- PMID: 39858813
- PMCID: PMC11767246
- DOI: 10.3390/microorganisms13010046
A Rewired NADPH-Dependent Redox Shuttle for Testing Peroxisomal Compartmentalization of Synthetic Metabolic Pathways in Komagataella phaffii
Abstract
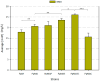
The introduction of heterologous pathways into microbial cell compartments offers several potential advantages, including increasing enzyme concentrations and reducing competition with native pathways, making this approach attractive for producing complex metabolites like fatty acids and fatty alcohols. However, measuring subcellular concentrations of these metabolites remains technically challenging. Here, we explored 3-hydroxypropionic acid (3-HP), readily quantifiable and sharing the same precursors-acetyl-CoA, NADPH, and ATP-with the above-mentioned products, as a reporter metabolite for peroxisomal engineering in the yeast Komagataella phaffii. To this end, the malonyl-CoA reductase pathway for 3-HP production was targeted into the peroxisome of K. phaffii using the PTS1-tagging system, and further tested with different carbon sources. Thereafter, we used compartmentalized 3-HP production as a reporter system to showcase the impact of different strategies aimed at enhancing the peroxisomal NADPH pool. Co-overexpression of genes encoding a NADPH-dependent redox shuttle from Saccharomyces cerevisiae (IDP2/IDP3) significantly increased 3-HP yields across all substrates, whereas peroxisomal targeting of the S. cerevisiae NADH kinase Pos5 failed to improve 3-HP production. This study highlights the potential of using peroxisomal 3-HP production as a biosensor for evaluating peroxisomal acetyl-CoA and NAPDH availability by simply quantifying 3-HP, demonstrating its potential for peroxisome-based metabolic engineering in yeast.
Keywords: 3-hydroxypropionic acid; Komagataella phaffii; NADPH; Pichia pastoris; acetyl-CoA; metabolic engineering; peroxisome; redox shuttle.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



References
-
- Faber K.N., Elgersma Y., Heyman J.A., Koller A., Lüers G.H., Nuttley W.M., Terlecky S.R., Wenzel T.J., Subramani S. Use of Pichia pastoris as a model eukaryotic system. Peroxisome biogenesis. Methods Mol. Biol. 1998;103:121–147. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

