Human Milk Archaea Associated with Neonatal Gut Colonization and Its Co-Occurrence with Bacteria
- PMID: 39858853
- PMCID: PMC11767358
- DOI: 10.3390/microorganisms13010085
Human Milk Archaea Associated with Neonatal Gut Colonization and Its Co-Occurrence with Bacteria
Abstract
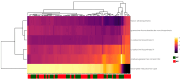
Archaea have been identified as early colonizers of the human intestine, appearing from the first days of life. It is hypothesized that the origin of many of these archaea is through vertical transmission during breastfeeding. In this study, we aimed to characterize the archaeal composition in samples of mother-neonate pairs to observe the potential vertical transmission. We performed a cross-sectional study characterizing the archaeal diversity of 40 human colostrum-neonatal stool samples by next-generation sequencing of V5-V6 16S rDNA libraries. Intra- and inter-sample analyses were carried out to describe the Archaeal diversity in each sample type. Human colostrum and neonatal stools presented similar core microbiota, mainly composed of the methanogens Methanoculleus and Methanosarcina. Beta diversity and metabolic prediction results suggest homogeneity between sample types. Further, the co-occurrence network analysis showed associations between Archaea and Bacteria, which might be relevant for these organisms' presence in the human milk and neonatal stool ecosystems. According to relative abundance proportions, beta diversity, and co-occurrence analyses, the similarities found imply that there is vertical transmission of archaea through breastfeeding. Nonetheless, differential abundances between the sample types suggest other relevant sources for colonizing archaea to the neonatal gut.
Keywords: 16S rDNA; Archaea; breastfeeding; human milk; microbiota; neonatal gut; vertical transmission.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures





References
-
- Pace R.M., Williams J.E., Robertson B., Lackey K.A., Meehan C.L., Price W.J., Foster J.A., Sellen D.W., Kamau-Mbuthia E.W., Kamundia E.W., et al. Variation in Human Milk Composition Is Related to Differences in Milk and Infant Fecal Microbial Communities. Microorganisms. 2021;9:1153. doi: 10.3390/microorganisms9061153. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

