De Novo Assembly of the Polyhydroxybutyrate (PHB) Producer Azohydromonas lata Strain H1 Genome and Genomic Analysis of PHB Production Machinery
- PMID: 39858905
- PMCID: PMC11767486
- DOI: 10.3390/microorganisms13010137
De Novo Assembly of the Polyhydroxybutyrate (PHB) Producer Azohydromonas lata Strain H1 Genome and Genomic Analysis of PHB Production Machinery
Abstract
Polyhydroxybutyrate (PHB) is a biodegradable natural polymer produced by different prokaryotes as a valuable carbon and energy storage compound. Its biosynthesis pathway requires the sole expression of the phaCAB operon, although auxiliary genes play a role in controlling polymer accumulation, degradation, granule formation and stabilization. Due to its biodegradability, PHB is currently regarded as a promising alternative to synthetic plastics for industrial/biotechnological applications. Azohydromonas lata strain H1 has been reported to accumulate PHB by using simple, inexpensive carbon sources. Here, we present the first de novo genome assembly of the A. lata strain H1. The genome assembly is over 7.7 Mb in size, including a circular megaplasmid of approximately 456 Kbp. In addition to the phaCAB operon, single genes ascribable to PhaC and PhaA functions and auxiliary genes were also detected. A comparative genomic analysis of the available genomes of the genus Azohydromonas revealed the presence of phaCAB and auxiliary genes in all Azohydromonas species investigated, suggesting that the PHB production is a common feature of the genus. Based on sequence identity, we also suggest A. australica as the closest species to which the phaCAB operon of the strain H1, reported in 1998, is similar.
Keywords: PHA; bacteria biopolymers; biodegradable; bioeconomy; bioplastic; polyhydroxyalkanoates.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
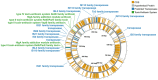

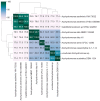
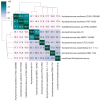


References
-
- Zaman A., Newman P. Plastics: Are they part of the zero-waste agenda or the toxic-waste agenda? Sustain. Earth. 2021;4:4. doi: 10.1186/s42055-021-00043-8. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

