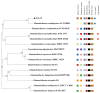
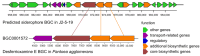
Association Analysis of the Genomic and Functional Characteristics of Halotolerant Glutamicibacter endophyticus J2-5-19 from the Rhizosphere of Suaeda salsa
- PMID: 39858975
- PMCID: PMC11767460
- DOI: 10.3390/microorganisms13010208
Association Analysis of the Genomic and Functional Characteristics of Halotolerant Glutamicibacter endophyticus J2-5-19 from the Rhizosphere of Suaeda salsa
Abstract
Halotolerant plant growth-promoting bacteria (HT-PGPB) have attracted considerable attention for their significant potential in mitigating salt stress in crops. However, the current exploration and development of HT-PGPB remain insufficient to meet the increasing demands of agriculture. In this study, an HT-PGPB isolated from coastal saline-alkali soil in the Yellow River Delta was identified as Glutamicibacter endophyticus J2-5-19. The strain was capable of growing in media with up to 13% NaCl and producing proteases, siderophores, and the plant hormone IAA. Under 4‱ salt stress, inoculation with strain J2-5-19 significantly increased the wheat seed germination rate from 37.5% to 95%, enhanced the dry weight of maize seedlings by 41.92%, and notably improved the development of maize root systems. Moreover, this work presented the first whole-genome of Glutamicibacter endophyticus, revealing that G. endophyticus J2-5-19 resisted salt stress by expelling sodium ions and taking up potassium ions through Na+/H+ antiporters and potassium uptake proteins, while also accumulating compatible solutes such as betaine, proline, and trehalose. Additionally, the genome contained multiple key plant growth-promoting genes, including those involved in IAA biosynthesis, siderophore production, and GABA synthesis. The findings provide a theoretical foundation and microbial resources for the development of specialized microbial inoculants for saline-alkali soils.
Keywords: Genomic analysis; Glutamicibacter endophyticus; Halotolerant plant growth-promoting bacteria; saline-alkali soil.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures










Similar articles
-
Halotolerant Enterobacter asburiae A103 isolated from the halophyte Salix linearistipularis: Genomic analysis and growth-promoting effects on Medicago sativa under alkali stress.Microbiol Res. 2024 Dec;289:127909. doi: 10.1016/j.micres.2024.127909. Epub 2024 Sep 19. Microbiol Res. 2024. PMID: 39305780
-
Growth-promoting bacteria and arbuscular mycorrhizal fungus enhance maize tolerance to saline stress.Microbiol Res. 2024 Jul;284:127708. doi: 10.1016/j.micres.2024.127708. Epub 2024 Apr 3. Microbiol Res. 2024. PMID: 38599021
-
[Microbial Diversity and Physicochemical Properties of Rhizosphere Microenvironment in Saline-alkali Soils of the Yellow River Delta].Huan Jing Ke Xue. 2020 Mar 8;41(3):1449-1455. doi: 10.13227/j.hjkx.201908044. Huan Jing Ke Xue. 2020. PMID: 32608648 Chinese.
-
Salt-tolerant plant growth-promoting bacteria as a versatile tool for combating salt stress in crop plants.Arch Microbiol. 2024 Jul 5;206(8):341. doi: 10.1007/s00203-024-04071-8. Arch Microbiol. 2024. PMID: 38967784 Review.
-
Salt-Tolerant Plant Growth-Promoting Bacteria (ST-PGPB): An Effective Strategy for Sustainable Food Production.Curr Microbiol. 2024 Aug 12;81(10):304. doi: 10.1007/s00284-024-03830-6. Curr Microbiol. 2024. PMID: 39133243 Review.
Cited by
-
Whole-Genome Analysis of Halomonas sp. H5 Revealed Multiple Functional Genes Relevant to Tomato Growth Promotion, Plant Salt Tolerance, and Rhizosphere Soil Microecology Regulation.Microorganisms. 2025 Jul 30;13(8):1781. doi: 10.3390/microorganisms13081781. Microorganisms. 2025. PMID: 40871285 Free PMC article.
References
-
- Singh A. Soil salinity: A global threat to sustainable development. Soil Use Manag. 2022;38:39–67. doi: 10.1111/sum.12772. - DOI
-
- Meena K.K., Bitla U., Sorty A.M., Kumar S., Kumar S., Wakchaure G.C., Singh D.P., Stougaard P., Suprasanna P. Understanding the Interaction and Potential of Halophytes and Associated Microbiome for Bio-saline Agriculture. J. Plant Growth Regul. 2023;42:6601–6619. doi: 10.1007/s00344-023-10912-5. - DOI
-
- Busse H.-J. Review of the taxonomy of the genus Arthrobacter, emendation of the genus Arthrobacter sensu lato, proposal to reclassify selected species of the genus Arthrobacter in the novel genera Glutamicibacter gen. nov., Paeniglutamicibacter gen. nov., Pseudoglutamicibacter gen. nov., Paenarthrobacter gen. nov. and Pseudarthrobacter gen. nov., and emended description of Arthrobacter roseus. Int. J. Syst. Evol. Microbiol. 2016;66:9–37. doi: 10.1099/ijsem.0.000702. - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

