High Antennal Expression of CYP6K1 and CYP4V2 Participate in the Recognition of Alarm Pheromones by Solenopsis invicta Buren
- PMID: 39859624
- PMCID: PMC11765799
- DOI: 10.3390/insects16010043
High Antennal Expression of CYP6K1 and CYP4V2 Participate in the Recognition of Alarm Pheromones by Solenopsis invicta Buren
Abstract
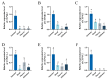
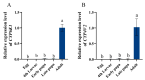
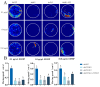
Insects have highly developed olfactory systems in which cytochrome P450s (CYPs) were involved as odor-degrading enzymes throughout the olfactory recognition of odor compounds by insects to avoid continuous stimulation of signaling molecules and thus damage to the olfactory nervous. To understand whether the highly expressed CYPs in the antennae play an olfactory function in Solenopsis invicta worker, in this study, we find six highly expressed antennal CYPs from the transcriptome of S. invicta. Multiple sequence alignment and phylogenetic analysis divided them into two families: the CYP3 family (SinvCYP6K1, SinvCYP6K1-1) and the CYP4 family (SinvCYP4C1, SinvCYP4C1-1, SinvCYP4C1-2, SinvCYP4V2). The expression patterns of these six CYPs were analyzed by RT-qPCR, which revealed that SinvCYP6K1 and SinvCYP4V2 were only highly expressed in the antennae of adult workers. The expression of SinvCYP6K1 and SinvCYP4V2 in workers was markedly diminished after feeding with dsRNA. The electroantennography (EAG) assay demonstrated that the silencing of either SinvCYP6K1 or SinvCYP4V2 resulted in a notable reduction in the EAG response of workers to 2-ethyl-3,6(5)-dimethylpyrazine (EDMP). Furthermore, the trajectory behavior assay showed that the worker's range and speed of movement in response to EDMP significant decreased after the silencing of SinvCYP6K1 and SinvCYP4V2. The findings indicated that both SinvCYP6K1 and SinvCYP4V2 were implicated in the recognition of EDMP by S. invicta.
Keywords: RNA interference; alarm pheromone; cytochrome P450s; electroantennography; red imported fire ant.
Conflict of interest statement
The authors declare that there are no conflicts of interest.
Figures



Similar articles
-
Functional Analysis of NPC2 in Alarm Pheromone Recognition by the Red Imported Fire Ant, Solenopsis invicta (Formicidae: Solenopsis).Insects. 2025 Jul 25;16(8):766. doi: 10.3390/insects16080766. Insects. 2025. PMID: 40870568 Free PMC article.
-
The Odorant Binding Protein, SiOBP5, Mediates Alarm Pheromone Olfactory Recognition in the Red Imported Fire Ant, Solenopsis invicta.Biomolecules. 2021 Oct 28;11(11):1595. doi: 10.3390/biom11111595. Biomolecules. 2021. PMID: 34827593 Free PMC article.
-
Electrophysiological and Alarm Responses of Solenopsis invicta Buren (Hymenoptera: Formicidae) to 2-Ethyl-3,5-dimethylpyrazine (Short Title: EAG and Behavioral Responses of Fire Ants to Pyrazine).Insects. 2019 Dec 13;10(12):451. doi: 10.3390/insects10120451. Insects. 2019. PMID: 31847156 Free PMC article.
-
HPLC Separation of 2-Ethyl-5(6)-methylpyrazine and Its Electroantennogram and Alarm Activities on Fire Ants (Solenopsis invicta Buren).Molecules. 2018 Jul 7;23(7):1661. doi: 10.3390/molecules23071661. Molecules. 2018. PMID: 29986521 Free PMC article.
-
Electroantennogram and behavioral responses of the imported fire ant, Solenopsis invicta Buren, to an alarm pheromone component and its analogues.J Agric Food Chem. 2014 Dec 10;62(49):11924-32. doi: 10.1021/jf505191s. Epub 2014 Nov 21. J Agric Food Chem. 2014. PMID: 25415443
Cited by
-
Functional Analysis of NPC2 in Alarm Pheromone Recognition by the Red Imported Fire Ant, Solenopsis invicta (Formicidae: Solenopsis).Insects. 2025 Jul 25;16(8):766. doi: 10.3390/insects16080766. Insects. 2025. PMID: 40870568 Free PMC article.
References
-
- Brito N.F., Moreira M.F., Melo A.C. A look inside odorant-binding proteins in insect chemoreception. J. Insect Physiol. 2016;95:51–65. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

