Unraveling TEOSINTE BRANCHED1/CYCLOIDEA/PROLIFERATING CELL FACTOR Transcription Factors in Safflower: A Blueprint for Stress Resilience and Metabolic Regulation
- PMID: 39860123
- PMCID: PMC11767934
- DOI: 10.3390/molecules30020254
Unraveling TEOSINTE BRANCHED1/CYCLOIDEA/PROLIFERATING CELL FACTOR Transcription Factors in Safflower: A Blueprint for Stress Resilience and Metabolic Regulation
Abstract
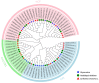
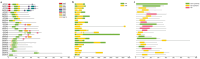
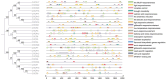
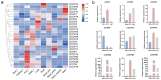
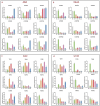
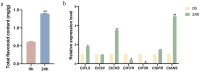
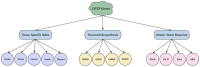
Safflower (Carthamus tinctorius L.), a versatile medicinal and economic crop, harbors untapped genetic resources essential for stress resilience and metabolic regulation. The TEOSINTE BRANCHED1/CYCLOIDEA/PROLIFERATING CELL FACTOR (TCP) transcription factors, exclusive to plants, are pivotal in orchestrating growth, development, and stress responses, yet their roles in safflower remain unexplored. Here, we report the comprehensive identification and characterization of 26 safflower TCP genes (CtTCPs), categorized into Class I (PROLIFERATING CELL FACTOR, PCF) and Class II (CINCINNATA and TEOSINTE BRANCHED1/CYCLOIDEA, CIN and CYC/TB1) subfamilies. Comparative phylogenetics, conserved motif, and gene structure analyses revealed a high degree of evolutionary conservation and functional divergence within the gene family. Promoter analyses uncovered light-, hormone-, and stress-responsive cis-elements, underscoring their regulatory potential. Functional insights from qRT-PCR analyses demonstrated dynamic CtTCP expression under abiotic stresses, including abscisic acid (ABA), Methyl Jasmonate (MeJA), Cold, and ultraviolet radiation b (UV-B) treatments. Notably, ABA stress triggered a significant increase in flavonoid accumulation, correlated with the upregulation of key flavonoid biosynthesis genes and select CtTCPs. These findings illuminate the complex regulatory networks underlying safflower's abiotic stress responses and secondary metabolism, offering a molecular framework to enhance crop resilience and metabolic engineering for sustainable agriculture.
Keywords: TCP transcription factors; flavonoid biosynthesis; gene expression profiling; molecular crop improvement; plant regulatory networks; safflower (Carthamus tinctorius L.).
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
Similar articles
-
Genome-wide identification, characterization and expression analysis of WRKY transcription factors under abiotic stresses in Carthamus tinctorius L.BMC Plant Biol. 2025 Jan 21;25(1):81. doi: 10.1186/s12870-025-06079-8. BMC Plant Biol. 2025. PMID: 39838282 Free PMC article.
-
Genome-wide identification and functional analyses of the TCP gene family in Carthamus tinctorius L.Sci Rep. 2025 Apr 15;15(1):12970. doi: 10.1038/s41598-025-97743-4. Sci Rep. 2025. PMID: 40234668 Free PMC article.
-
Genome-Wide Identification and Expression Analysis of TCP Transcription Factors Responding to Multiple Stresses in Arachis hypogaea L.Int J Mol Sci. 2025 Jan 26;26(3):1069. doi: 10.3390/ijms26031069. Int J Mol Sci. 2025. PMID: 39940846 Free PMC article.
-
The Regulation of CIN-like TCP Transcription Factors.Int J Mol Sci. 2020 Jun 24;21(12):4498. doi: 10.3390/ijms21124498. Int J Mol Sci. 2020. PMID: 32599902 Free PMC article. Review.
-
TCP Transcription Factors in Plant Reproductive Development: Juggling Multiple Roles.Biomolecules. 2023 Apr 26;13(5):750. doi: 10.3390/biom13050750. Biomolecules. 2023. PMID: 37238620 Free PMC article. Review.
References
-
- Kutsenkova V.S., Nepovinnykh N.V., Guo Q.B. Using of safflower seeds as a protein fortifier for shortbread. Food Hydrocoll. 2020;105:105808.
-
- Deviren H., Aydin H. Production and physicochemical properties of safflower seed oil extracted using different methods and its conversion to biodiesel. Fuel. 2023;343:128001. doi: 10.1016/j.fuel.2023.128001. - DOI
-
- Do K.L., Mushtaq A., Ahsan T., Yousaf M., Zhao F., Su M. Flavonoid-based yellow dye extract from safflower (Carthamus tinctorius L.) combined with chitosan for anti-bacterial and ultraviolet-protective functionalisation of silk. Color. Technol. 2024;140:900–912. doi: 10.1111/cote.12750. - DOI
MeSH terms
Substances
Grants and funding
- No. 20220204058YY, 20220505019ZP/the Science and Technology Development Project of Jilin Province
- No.31771868/Xinjiang safflower industry development fund,the National Natural Science Foundation of China
- No.JJKH20230400KJ/Science and Technology Research Program of Education Department of Jilin province
LinkOut - more resources
Full Text Sources

