First Report of SNPs Detection in TMEM154 Gene in Sheep from Poland and Their Association with SRLV Infection Status
- PMID: 39860977
- PMCID: PMC11768335
- DOI: 10.3390/pathogens14010016
First Report of SNPs Detection in TMEM154 Gene in Sheep from Poland and Their Association with SRLV Infection Status
Abstract
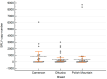
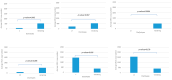
Small ruminant lentiviruses (SRLVs) infect sheep, causing a multiorganic disease called maedi-visna or ovine progressive pneumonia, which significantly affects the production and welfare of sheep, generating serious economic losses. Although not all infected animals develop fully symptomatic disease, they constantly spread the virus in the flock. Since the infection is incurable and no vaccine is available, another approach is necessary to control SRLV infections. In recent years, an alternative for culling infected animals has become the approach based on identifying genetic markers for selecting SRLV-resistant individuals. Recent reports revealed several candidates, including gene encoding transmembrane protein 154 (TMEM154). Several single nucleotide polymorphisms (SNPs) are found within this gene in sheep of different breeds, but only some can be considered as resistant markers. This study aimed to investigate the presence of single polymorphic sites in TMEM154 gene in sheep of selected Polish flocks and assess their association with the infection and proviral load in the context of susceptibility to SRLV infection. In total 107 sheep, representing three breeds, were screened for their SRLV infection status by serological and PCR testing. All these animals were also genotyped to characterize the presence of SNPs in TMEM154 gene and estimate their potential of being the SRLV-resistance marker. The frequency of identified alleles differed among breeds. Moreover, the positive association between TMEM154 genotype and SRLV status was found for E35K polymorphism and two polymorphic sites in 5'UTR in one of analyzed breed. However, when the relationship between SNPs and SRLV proviral load was analyzed, five had a strong association, considering the whole population of tested sheep. Presented data, for the first time, identified the presence of SNPs in TMEM154 gene in sheep housed in Polish flocks and suggested that selecting SRLV-resistant animals based on this analysis might be possible, but further validation in a larger group of sheep is required.
Keywords: TMEM154; genotyping; sheep; single nucleotide polymorphism; small ruminant lentiviruses (SRLVs).
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

Similar articles
-
Small ruminant lentivirus genetic subgroups associate with sheep TMEM154 genotypes.Vet Res. 2013 Jul 29;44(1):64. doi: 10.1186/1297-9716-44-64. Vet Res. 2013. PMID: 23895262 Free PMC article.
-
Mutations in Ovis aries TMEM154 are associated with lower small ruminant lentivirus proviral concentration in one sheep flock.Anim Genet. 2014 Aug;45(4):565-71. doi: 10.1111/age.12181. Epub 2014 Jun 17. Anim Genet. 2014. PMID: 24934128 Free PMC article.
-
Identification of New Single Nucleotide Polymorphisms Potentially Related to Small Ruminant Lentivirus Infection Susceptibility in Goats Based on Data Selected from High-Throughput Sequencing.Pathogens. 2024 Sep 25;13(10):830. doi: 10.3390/pathogens13100830. Pathogens. 2024. PMID: 39452702 Free PMC article.
-
Progress on ovine lentivirus and its resistant genes.Yi Chuan. 2014 Dec;36(12):1204-10. doi: 10.3724/SP.J.1005.2014.1204. Yi Chuan. 2014. PMID: 25487264 Review.
-
Small Ruminant Lentiviruses and Caseous Lymphadenitis.Vet Clin North Am Food Anim Pract. 2025 Mar;41(1):83-92. doi: 10.1016/j.cvfa.2024.11.005. Epub 2024 Nov 28. Vet Clin North Am Food Anim Pract. 2025. PMID: 39609153 Review.
References
-
- Peterhans E., Greenland T., Badiola J., Harkiss G., Bertoni G., Amorena B., Eliaszewicz M., Juste R.A., Krassnig R., Lafont J.-P., et al. Routes of transmission and consequences of small ruminant lentiviruses (SRLVs) infection and eradication schemes. Vet. Res. 2004;35:257–274. doi: 10.1051/vetres:2004014. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

