Mitochondrial DNA Structure in Trypanosoma cruzi
- PMID: 39861034
- PMCID: PMC11769408
- DOI: 10.3390/pathogens14010073
Mitochondrial DNA Structure in Trypanosoma cruzi
Abstract
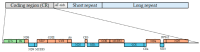
Kinetoplastids display a single, large mitochondrion per cell, with their mitochondrial DNA referred to as the kinetoplast. This kinetoplast is a network of concatenated circular molecules comprising a maxicircle (20-64 kb) and up to thousands of minicircles varying in size depending on the species (0.5-10 kb). In Trypanosoma cruzi, maxicircles contain typical mitochondrial genes found in other eukaryotes. They consist of coding and divergent/variable regions, complicating their assembly due to repetitive elements. However, next-generation sequencing (NGS) methods have resolved these issues, enabling the complete sequencing of maxicircles from different strains. Furthermore, several insertions and deletions in the maxicircle sequences have been identified among strains, affecting specific genes. Unique to kinetoplastids, minicircles play a crucial role in a particular U-insertion/deletion RNA editing system by encoding guide RNAs (gRNAs). These gRNAs are essential for editing and maturing maxicircle mRNAs. In Trypanosoma cruzi, although only a few studies have utilized NGS methods to date, the structure of these molecules suggests a classification into four main groups of minicircles. This classification is based on their size and the number of highly conserved regions (mHCRs) and hypervariable regions (mHVRs).
Keywords: Trypanosoma cruzi; kinetoplast; maxicircles; minicircles.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

Similar articles
-
The Complete Mitochondrial DNA of Trypanosoma cruzi: Maxicircles and Minicircles.Front Cell Infect Microbiol. 2021 Jun 29;11:672448. doi: 10.3389/fcimb.2021.672448. eCollection 2021. Front Cell Infect Microbiol. 2021. PMID: 34268138 Free PMC article.
-
A population study of the minicircles in Trypanosoma cruzi: predicting guide RNAs in the absence of empirical RNA editing.BMC Genomics. 2007 May 24;8:133. doi: 10.1186/1471-2164-8-133. BMC Genomics. 2007. PMID: 17524149 Free PMC article.
-
Trypanosoma cruzi mitochondrial maxicircles display species- and strain-specific variation and a conserved element in the non-coding region.BMC Genomics. 2006 Mar 22;7:60. doi: 10.1186/1471-2164-7-60. BMC Genomics. 2006. PMID: 16553959 Free PMC article.
-
Mitochondrial RNA editing in trypanosomes: small RNAs in control.Biochimie. 2014 May;100:125-31. doi: 10.1016/j.biochi.2014.01.003. Epub 2014 Jan 17. Biochimie. 2014. PMID: 24440637 Free PMC article. Review.
-
The structure and replication of kinetoplast DNA.Annu Rev Microbiol. 1995;49:117-43. doi: 10.1146/annurev.mi.49.100195.001001. Annu Rev Microbiol. 1995. PMID: 8561456 Review.
Cited by
-
TrypPROTACs Unlocking New Therapeutic Strategies for Chagas Disease.Pharmaceuticals (Basel). 2025 Jun 19;18(6):919. doi: 10.3390/ph18060919. Pharmaceuticals (Basel). 2025. PMID: 40573314 Free PMC article. Review.
References
-
- Zingales B., Miles M.A., Campbell D.A., Tibayrenc M., Macedo A.M., Teixeira M.M.G., Schijman A.G., Llewellyn M.S., Lages-Silva E., Machado C.R., et al. The Revised Trypanosoma Cruzi Subspecific Nomenclature: Rationale, Epidemiological Relevance and Research Applications. Infect. Genet. Evol. J. Mol. Epidemiol. Evol. Genet. Infect. Dis. 2012;12:240–253. doi: 10.1016/j.meegid.2011.12.009. - DOI - PubMed
-
- Barnabé C., Mobarec H.I., Jurado M.R., Cortez J.A., Brenière S.F. Reconsideration of the Seven Discrete Typing Units within the Species Trypanosoma Cruzi, a New Proposal of Three Reliable Mitochondrial Clades. Infect. Genet. Evol. J. Mol. Epidemiol. Evol. Genet. Infect. Dis. 2016;39:176–186. doi: 10.1016/j.meegid.2016.01.029. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

