Τhiazolidine-4-One Derivatives with Variable Modes of Inhibitory Action Against DPP4, a Drug Target with Multiple Activities and Established Role in Diabetes Mellitus Type II
- PMID: 39861115
- PMCID: PMC11768251
- DOI: 10.3390/ph18010052
Τhiazolidine-4-One Derivatives with Variable Modes of Inhibitory Action Against DPP4, a Drug Target with Multiple Activities and Established Role in Diabetes Mellitus Type II
Abstract
Background/Objectives: DPP4 is an enzyme with multiple natural substrates and probable involvement in various mechanisms. It constitutes a drug target for the treatment of diabetes II, although, also related to other disorders. While a number of drugs with competitive inhibitory action and covalent binding capacity are available, undesired side effects exist partly attributed to drug kinetics, and research for finding novel, potent, and safer compounds continues. Despite the research, a low number of uncompetitive and non-competitive inhibitors, which could be of worth for pharmaceutical and mechanism studies, was mentioned. Methods: In the present study sixteen 3-(benzo[d]thiazol-2-yl)-2-aryl thiazolidin-4-ones were selected for evaluation, based on structural characteristics and docking analysis and were tested in vitro for DPP4 inhibitory action using H-Gly-Pro-amidomethyl coumarin substrate. Their mode of inhibition was also in vitro explored. Results: Twelve compounds exhibited IC50 values at the nM range with the best showing IC50 = 12 ± 0.5 nM, better than sitagliptin. Most compounds exhibited a competitive mode of inhibition. Inhibition modes of uncompetitive, non-competitive, and mixed type were also identified. Docking analysis was in accordance with the in vitro results, with a linear correlation of logIC50 with a Probability of Binding Factor(PF) derived using docking analysis to a specific target box and to the whole enzyme. According to the docking results, two probable sites of binding for uncompetitive inhibitors were highlighted in the wider area of the active site and in the propeller loop. Conclusions: Potent inhibitors with IC50 at the nM range and competitive, non-competitive, uncompetitive, and mixed modes of action, one better than sitagliptin, were found. Docking analysis was used to estimate probable sites and ways of binding. However, crystallographic or NMR studies are needed to elucidate the exact way of binding especially for uncompetitive and non-competitive inhibitors.
Keywords: 3-(benzo[d]thiazol-2-yl)-2-aryl thiazolidin-4-ones; DPP4; allosteric center; competitive inhibitors; diabetes mellitus type II; docking analysis; mixed inhibition; non-competitive inhibitors; probability factor for binding PF; uncompetitive inhibitors.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


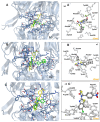


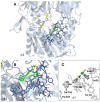
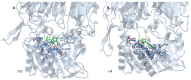




References
-
- Frayling T.M., Evans J.C., Bulman M.P., Pearson E., Allen L., Owen K., Bingham C., Hannemann M., Shepherd M., Ellard S., et al. Beta-Cell Genes and Diabetes: Molecular and Clinical Characterization of Mutations in Transcription Factors. Diabetes. 2001;50((Suppl. 1)):S94. doi: 10.2337/diabetes.50.2007.S94. - DOI - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

