Genome-Wide Identification and Expression Analysis of bHLH-MYC Family Genes from Mustard That May Be Important in Trichome Formation
- PMID: 39861625
- PMCID: PMC11769027
- DOI: 10.3390/plants14020268
Genome-Wide Identification and Expression Analysis of bHLH-MYC Family Genes from Mustard That May Be Important in Trichome Formation
Abstract
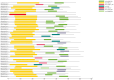
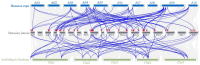
The trichomes of mustard leaves have significance due to their ability to combat unfavorable external conditions and enhance disease resistance. It was demonstrated that the MYB-bHLH-WD40 (MBW) ternary complex consists of MYB, basic Helix-Loop-Helix (bHLH), and WD40-repeat (WD40) family proteins and plays a key role in regulating trichome formation and density. The bHLH gene family, particularly the Myelocytomatosis (MYC) proteins that possess the structural bHLH domain (termed bHLH-MYC), are crucial to the formation and development of leaf trichomes in plants. bHLH constitutes one of the largest families of transcription factors in eukaryotes, of which MYC is a subfamily member. However, studies on bHLH-MYC transcription factors in mustard have yet to be reported. In this study, a total of 45 bHLH-MYC transcription factors were identified within the Brassica juncea genome, and a comprehensive series of bioinformatic analyses were conducted on their structures and properties: an examination of protein physicochemical properties, an exploration of conserved structural domains, an assessment of chromosomal positional distributions, an analysis of the conserved motifs, an evaluation of the gene structures, microsynteny analyses, three-dimensional structure prediction, and an analysis of sequence signatures. Finally, transcriptome analyses and a subcellular localization examination were performed. The results revealed that these transcription factors were unevenly distributed across 18 chromosomes, showing relatively consistent conserved motifs and gene structures and high homology. The final results of the transcriptome analysis and gene annotation showed a high degree of variability in the expression of bHLH-MYC transcription factors. Five genes that may be associated with trichome development (BjuVA09G22490, BjuVA09G13750, BjuVB04G14560, BjuVA05G24810, and BjuVA06G44820) were identified. The subcellular localization results indicated that the transcription and translation products of these five genes were expressed in the same organelle: the nucleus. This finding provides a basis for elucidating the roles of bHLH-MYC family members in plant growth and development, and the molecular mechanisms underlying trichome development in mustard leaves.
Keywords: MBW; MYC; bHLH; leaf trichome; mustard.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures








Similar articles
-
SlMYB72 and SlMYB75 antagonistically regulate trichome formation via the MYB-bHLH-WD40 complex in tomato.J Biol Chem. 2025 Mar;301(3):108313. doi: 10.1016/j.jbc.2025.108313. Epub 2025 Feb 13. J Biol Chem. 2025. PMID: 39955063 Free PMC article.
-
Genome-Wide Identification of Direct Targets of the TTG1-bHLH-MYB Complex in Regulating Trichome Formation and Flavonoid Accumulation in Arabidopsis Thaliana.Int J Mol Sci. 2019 Oct 10;20(20):5014. doi: 10.3390/ijms20205014. Int J Mol Sci. 2019. PMID: 31658678 Free PMC article.
-
Characterization and expression analysis of GLABRA3 (GL3) genes in cotton: insights into trichome development and hormonal regulation.Mol Biol Rep. 2024 Apr 5;51(1):479. doi: 10.1007/s11033-024-09412-w. Mol Biol Rep. 2024. PMID: 38578511
-
An overview of the gene regulatory network controlling trichome development in the model plant, Arabidopsis.Front Plant Sci. 2014 Jun 5;5:259. doi: 10.3389/fpls.2014.00259. eCollection 2014. Front Plant Sci. 2014. PMID: 25018756 Free PMC article. Review.
-
Evolutionary studies of the bHLH transcription factors belonging to MBW complex: their role in seed development.Ann Bot. 2023 Nov 23;132(3):383-400. doi: 10.1093/aob/mcad097. Ann Bot. 2023. PMID: 37467144 Free PMC article. Review.
Cited by
-
Bioinformatics analysis and expression of the transcription factor MYC2 in Brassica napus in response to salt stress.Funct Integr Genomics. 2025 Jul 1;25(1):141. doi: 10.1007/s10142-025-01647-5. Funct Integr Genomics. 2025. PMID: 40591005
References
-
- Chen C.L., Zhou G.F., Fan Y.H., Zhou Y., Chen X.Q. Discussion on the origin of mustard (Brassica juncea) in china. Acta Horticulturae. 1995;402:317–320.
-
- Kim E.J., Choi J.Y., Yu M.R., Kim M.Y., Lee S.H., Lee B.H. Total polyphenols, total flavonoid contents, and antioxidant activity of Korean natural and medicinal plants. Korean J. Food Sci. Technol. 2012;44:337–342. doi: 10.9721/KJFST.2012.44.3.337. - DOI
LinkOut - more resources
Full Text Sources

