Identifying Allosteric Small-Molecule Binding Sites of Inactive NS2B-NS3 Proteases of Pathogenic Flaviviridae
- PMID: 39861795
- PMCID: PMC11769402
- DOI: 10.3390/v17010006
Identifying Allosteric Small-Molecule Binding Sites of Inactive NS2B-NS3 Proteases of Pathogenic Flaviviridae
Abstract
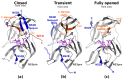
Dengue, West Nile, Zika, Yellow fever, and Japanese encephalitis viruses persist as significant global health threats. The development of new therapeutic strategies based on inhibiting essential viral enzymes or viral-host protein interactions is problematic due to the fast mutation rate and rapid emergence of drug resistance. This study focuses on the NS2B-NS3 protease as a promising target for antiviral drug development. Promising allosteric binding sites were identified in two conformationally distinct inactive states and characterized for five flaviviruses and four Dengue virus subtypes. Their shapes, druggability, inter-viral similarity, sequence variation, and susceptibility to drug-resistant mutations have been studied. Two identified allosteric inactive state pockets appear to be feasible alternatives to a larger closed pocket near the active site, and they can be targeted with specific drug-like small-molecule inhibitors. Virus-specific sequence and structure implications and the feasibility of multi-viral inhibitors are discussed.
Keywords: Dengue; Japanese encephalitis; NS2B; NS3; Yellow Fever; Zika virus; allosteric druggable pockets; mutation rates; protease inhibitors.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures





References
-
- CDC About Viral Hemorrhagic Fevers. [(accessed on 17 November 2024)]; Available online: https://www.cdc.gov/viral-hemorrhagic-fevers/about/index.html.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

