Atlas of Interactions Between Decoration Proteins and Major Capsid Proteins of Coliphage N4
- PMID: 39861808
- PMCID: PMC11768535
- DOI: 10.3390/v17010019
Atlas of Interactions Between Decoration Proteins and Major Capsid Proteins of Coliphage N4
Abstract
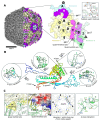
Coliphage N4 is a representative species of the Schitoviridae family of bacteriophages. Originally structurally studied in 2008, the capsid structure was solved to 14 Å to reveal an interesting arrangement of Ig-like decoration proteins across the surface of the capsid. Herein, we present a high-resolution N4 structure, reporting a 2.45 Å map of the capsid obtained via single particle cryogenic-electron microscopy. Structural analysis of the major capsid proteins (MCPs) and decoration proteins (gp56 and gp17) of phage N4 reveals a pattern of interactions across the capsid that are mediated by structurally homologous domains of gp17. In this study, an analysis of the complex interface contacts allows us to confirm that the gp17 Ig-like decoration proteins of N4 are likely employed by the virus to increase the capsid's structural integrity.
Keywords: Ig-like decoration protein; bacteriophage N4; capsid decoration protein; cryo-electron microscopy.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

Similar articles
-
About bacteriophage tail terminator and tail completion proteins: structure of the proximal extremity of siphophage T5 tail.J Virol. 2025 Jan 31;99(1):e0137624. doi: 10.1128/jvi.01376-24. Epub 2024 Dec 23. J Virol. 2025. PMID: 39714170 Free PMC article.
-
Murine norovirus allosteric escape mutants mimic gut activation.J Virol. 2025 Jun 17;99(6):e0021925. doi: 10.1128/jvi.00219-25. Epub 2025 May 12. J Virol. 2025. PMID: 40353669 Free PMC article.
-
Templated trimerization of the phage L decoration protein on capsids.Protein Sci. 2025 Apr;34(4):e70089. doi: 10.1002/pro.70089. Protein Sci. 2025. PMID: 40100157
-
Structural Capsidomics of Single-Stranded DNA Viruses.Viruses. 2025 Feb 27;17(3):333. doi: 10.3390/v17030333. Viruses. 2025. PMID: 40143263 Free PMC article. Review.
-
Emerging structure of chlorovirus PBCV-1.Virology. 2025 Jul;608:110552. doi: 10.1016/j.virol.2025.110552. Epub 2025 Apr 22. Virology. 2025. PMID: 40286469 Review.
References
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources

