Virological Aspects of COVID-19 in Patients with Hematological Malignancies: Duration of Viral Shedding and Genetic Analysis
- PMID: 39861838
- PMCID: PMC11768452
- DOI: 10.3390/v17010046
Virological Aspects of COVID-19 in Patients with Hematological Malignancies: Duration of Viral Shedding and Genetic Analysis
Abstract
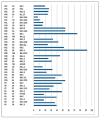
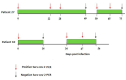
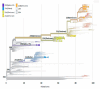
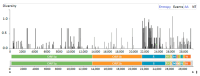
Coronavirus disease 2019 (COVID-19) has been associated with a significant fatality rate and persistent evolution in immunocompromised patients. In this prospective study, we aimed to determine the duration of excretion of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in 37 Tunisian patients with hematological malignancies (40.5% with lymphoma and 37.8% with leukemia). In order to investigate the accumulation of viral mutations, we carried out genetic investigation on longitudinal nasopharyngeal samples using RT-PCR and whole-genome sequencing. Patients' samples were collected until the RT-PCR results became negative. SARS-CoV-2 infection was symptomatic in 48.6% of cases with fever, and cough was symptomatic in 61% of cases; the mortality rate was estimated to be 13.5%. The duration of viral RNA shedding ranged from 7 to 92 days after onset; it exceeded 18 days in 79.4% of cases. An intermittent PCR positivity was observed in two symptomatic patients. Persistent PCR positivity, defined as the presence of viral RNA for more than 30 days, was found in 51.4% of cases. No significant differences were observed for age, sex, type of hematological malignancy, or COVID-19 evolution between this group and a second one characterized by non-persistent PCR positivity. Lymphopenia was an independent predictor of prolonged SARS-CoV-2 RNA detection (p = 0.04). Three types of variants were detected; the most frequent was the Omicron. Globally, the mean intra-host variability in the SARS-CoV-2 genome was 1.31 × 10-3 mutations per site per year; it was 1.44 × 10-3 in the persistent group and 1.3 × 10-3 in the non-persistent group. Three types of mutations were detected; the most frequent were nucleotide substitutions in the spike (S) gene. No statistically significant difference was observed between the two groups as to the type and mean number of observed mutations in the whole genome and the S region (p = 0.650). Sequence analysis revealed the inclusion of one to eight amino acid-changing events in seventeen cases; it was characterized by genetic stability from the third to the twentieth day of evolution in six cases. For the two patients with intermittent PCR positivity, sequences obtained from samples before and after negative PCR were identical in the whole genome, confirming an intra-host evolution of the same viral strain. This study confirms the risk of persistent viral shedding in patients with hematological malignancies. However, persistence of PCR positivity seems to be correlated only with a continuous elimination of viral RNA debris. Additional studies based on cell culture analysis are needed to confirm these findings.
Keywords: SARS-CoV-2 infection; hematological malignancies; viral shedding; whole-genome sequencing.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Aleem A., Akbar Samad A.B., Vaqar S. StatPearls. StatPearls; Treasure Island, FL, USA: 2024. Emerging Variants of SARS-CoV-2 and Novel Therapeutics Against Coronavirus (COVID-19) Bookshelf ID: NBK570580. - PubMed
-
- Papanikolaou V., Chrysovergis A., Ragos V., Tsiambas E., Katsinis S., Manoli A., Papouliakos S., Roukas D., Mastronikolis S., Peschos D., et al. From delta to Omicron: S1-RBD/S2 mutation/deletion equilibrium in SARS-CoV-2 defined variants. Gene. 2022;814:146134. doi: 10.1016/j.gene.2021.146134. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

