The Hao-Fountain syndrome protein USP7 regulates neuronal connectivity in the brain via a novel p53-independent ubiquitin signaling pathway
- PMID: 39862434
- PMCID: PMC11922642
- DOI: 10.1016/j.celrep.2025.115231
The Hao-Fountain syndrome protein USP7 regulates neuronal connectivity in the brain via a novel p53-independent ubiquitin signaling pathway
Abstract
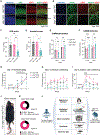
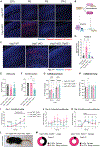
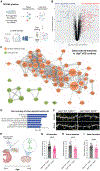
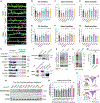
Mutation or deletion of the deubiquitinase USP7 causes Hao-Fountain syndrome (HAFOUS), which is characterized by speech delay, intellectual disability, and aggressive behavior and highlights important unknown roles of USP7 in the nervous system. Here, we conditionally delete USP7 in glutamatergic neurons in the mouse forebrain, triggering disease-relevant phenotypes, including sensorimotor deficits, impaired cognition, and aggressive behavior. Although USP7 deletion induces p53-dependent neuronal apoptosis, most behavioral abnormalities in USP7 conditional knockout mice persist following p53 loss. Strikingly, USP7 deletion perturbs the synaptic proteome and dendritic spinogenesis independent of p53. Integrated proteomics and biochemical analyses identify the RNA splicing factor Ppil4 as a key substrate of USP7. Ppil4 knockdown phenocopies the effect of USP7 loss on dendritic spines. Accordingly, USP7 loss disrupts splicing of synaptic genes. These findings reveal that USP7-Ppil4 signaling regulates neuronal connectivity in the developing brain with implications for our understanding of HAFOUS pathogenesis and other neurodevelopmental disorders.
Keywords: CP: Molecular biology; CP: Neuroscience; HAFOUS; Hao-Fountain syndrome; Ppil4; RNA splicing; TMT proteomics; USP7; brain development; deubiquitinase; p53; synapse; ubiquitin.
Copyright © 2025 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests A.H.K. is a consultant for Monteris Medical and has received a research grant from Stryker to study a dural substitute, both of which have no direct relation to this study. A.B. is a full-time employee and shareholder of F. Hoffmann-La Roche Ltd.
Figures



Update of
-
The Hao-Fountain syndrome protein USP7 regulates neuronal connectivity in the brain via a novel p53-independent ubiquitin signaling pathway.bioRxiv [Preprint]. 2024 Jun 29:2023.10.24.563880. doi: 10.1101/2023.10.24.563880. bioRxiv. 2024. Update in: Cell Rep. 2025 Feb 25;44(2):115231. doi: 10.1016/j.celrep.2025.115231. PMID: 37961719 Free PMC article. Updated. Preprint.
References
-
- Ferguson CJ, Urso O, Bodrug T, Gassaway BM, Watson ER, Prabu JR, Lara-Gonzalez P, Martinez-Chacin RC, Wu DY, Brigatti KW, et al. (2022). APC7 mediates ubiquitin signaling in constitutive heterochromatin in the developing mammalian brain. Mol. Cell 82, 90–105.e13. 10.1016/j.molcel.2021.11.031. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

