Topologically constrained DNA-mediated one-pot CRISPR assay for rapid detection of viral RNA with single nucleotide resolution
- PMID: 39862805
- PMCID: PMC11873568
- DOI: 10.1016/j.ebiom.2025.105564
Topologically constrained DNA-mediated one-pot CRISPR assay for rapid detection of viral RNA with single nucleotide resolution
Abstract
Background: The widespread and evolution of RNA viruses, such as the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), highlights the importance of fast identification of virus subtypes, particularly in non-laboratory settings. Rapid and inexpensive at-home testing of viral nucleic acids with single-base resolution remains a challenge.
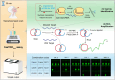
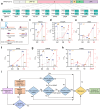
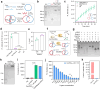
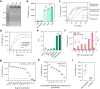
Methods: Topologically constrained DNA ring is engineered as substrates for the trans-cleavage of Cas13a to yield an accelerated post isothermal amplification. The capacity of CRISPR/Cas13a for discriminating single nucleotide variant (SNV) in viral genome is leveraged by designing synthetic mismatches and hairpin structure in CRISPR RNA (crRNA), enabling robust discrimination of different SARS-CoV-2 variants. Via optimisation of CasTDR3pot to be one-pot assay, CasTDR1pot can detect Omicron and its subvariants, with only a few copies in clinical samples in less than 30 min without pre-amplification.
Findings: The detection system boasts high sensitivity (0.1 aM), single-base specificity, and the advantage of a rapid "sample-to-answer" process, which takes only 30 min. In the detection of SARS-CoV-2 clinical samples and their variant strains, CasTDR1pot has achieved 100% accuracy. Furthermore, the design of a portable signal-reading device facilitates user-friendly result interpretation. For the detection needs of different RNA viruses, the system can be adapted simply by designing the corresponding crRNA.
Interpretation: Our study provides a rapid and accurate molecular diagnostic tool for point-of-care testing, epidemiological screening, and the detection of diseases associated with other RNA biomarkers with excellent single nucleotide differentiation, high sensitivity, and simplicity.
Funding: National Key Research and Development Program of China (No. 2023YFB3208302), National Natural Science Foundation of China (No. 22377110, 22034004, 82402749, 82073787, 22122409), National Key Research and Development Program of China (No. 2021YFA1200104), Henan Province Fund for Cultivating Advantageous Disciplines (No. 222301420019).
Keywords: CRISPR diagnostic; One-pot assay; SARS-CoV-2 variants; Single base mutation; Topologically constrained DNA rings.
Copyright © 2025 The Author(s). Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interest.
Figures


References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

