A Primer on Proteomic Characterization of Intercellular Communication in a Virus Microenvironment
- PMID: 39862905
- PMCID: PMC11889360
- DOI: 10.1016/j.mcpro.2025.100913
A Primer on Proteomic Characterization of Intercellular Communication in a Virus Microenvironment
Abstract
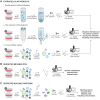
Intercellular communication is fundamental to multicellular life and a core determinant of outcomes during viral infection, where the common goals of virus and host for persistence and replication are generally at odds. Hosts rely on encoded innate and adaptive immune responses to detect and clear viral pathogens, while viruses can exploit or disrupt these pathways and other intercellular communication processes to enhance their spread and promote pathogenesis. While virus-induced signaling can result in systemic changes to the host, striking alterations are observed within the cellular microenvironment directly surrounding a site of infection, termed the virus microenvironment (VME). Mechanisms employed by viruses to condition their VMEs are emerging and are critical for understanding the biology and pathologies of viral infections. Recent advances in experimental approaches, including proteomic methods, have enabled study of the VME in unprecedented detail. In this review article, we provide a primer on proteomic approaches used to study how viral infections alter intercellular communication, highlighting the ways in which these approaches have been implemented and the exciting biology they have uncovered. First, we consider the different molecules secreted by an infected cell, including proteins, either soluble or contained within extracellular vesicles, and metabolites. We further discuss the modalities of interactions facilitated by alteration at the cell surface of infected cells, including immunopeptide presentation and interactions with the extracellular matrix. Finally, we review spatial profiling approaches that have allowed distinguishing how specific subpopulations of cells within a VME respond to infection and alter their protein composition, discussing valuable insights these methods have offered.
Keywords: cell surface; cell-to-cell communication; extracellular matrix; extracellular vesicles; intercellular communication; microenvironment labeling; multiplexed imaging; proteomics; secreted metabolites; secreted proteins; secretome; spatial profiling; surfaceome; viral infection; virus microenvironment.
Copyright © 2025 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflicts of Interest The authors declare no competing interests.
Figures




References
-
- Carty M., Guy C., Bowie A.G. Detection of viral infections by innate immunity. Biochem. Pharmacol. 2021;183 - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

